3 a3 Format
The Amino Acid Annotation (a3) format is created to hold amino acid sequences and annotations.
An example of the a3 format is shown below:
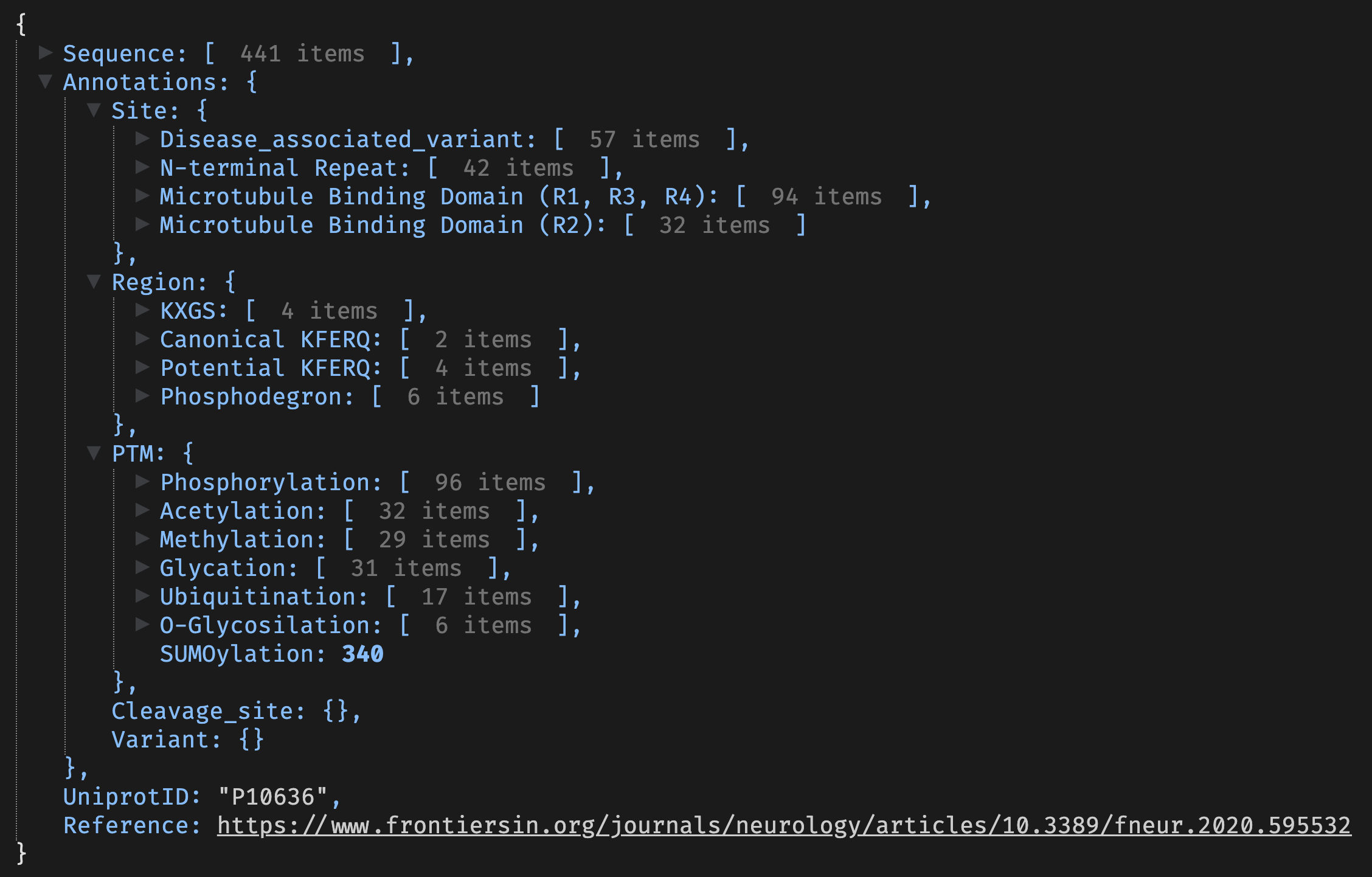
a3 format example showing the nested list structure of annotations.It is a nested list of lists. At the top level, there are:
- Sequence A character vector containing the amino acid sequence.
- Annotations A named list of lists containing different types of annotations.
- UniprotID A character containing the Uniprot ID of the protein if available.
- Reference A URL of a relevant publication if available.
3.1 Annotations
There are 5 different annotation types defined in the a3 format, and more can be added as needed.
- Site A named list of (unsigned) integer vectors. The name gives the type of annotation, and the vector contains the amino acid positions of the annotation.
- Region A named list of integer vectors. The name gives the type of annotation, and the vector contains the start and end positions of the annotation.
- PTM A named list of integer vectors with positions for each PTM.
- Cleavage Sites A named list of integer vectors with positions for each cleavage site.
- Variant A list of lists with variant information. Each element list can contain any number of named elements for the given variant.
