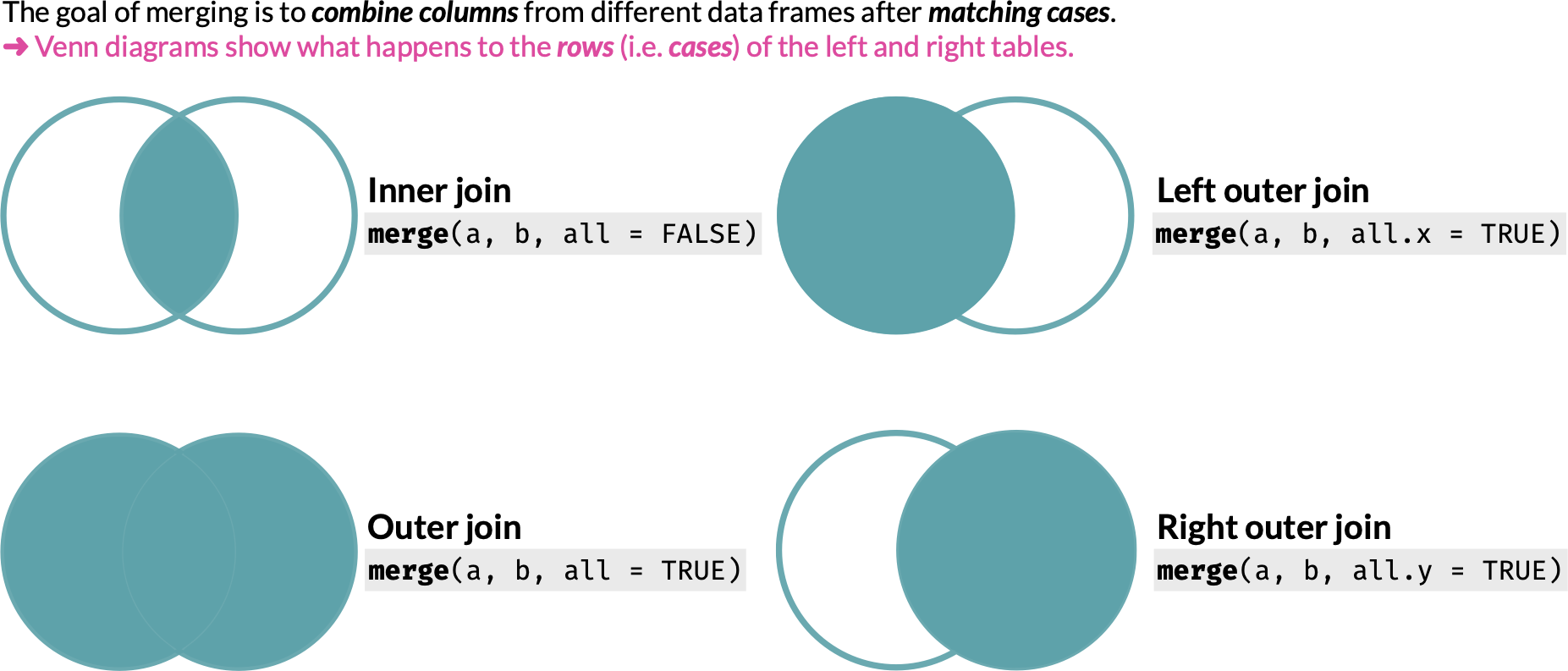
27 Joins
We often have data from separate sources that we want to combine into a single data.frame. Table joins allow you to specify how to perform such a merge.
There are four main types of join operations:

Scenario: You have received two tables with clinical data. Each table contains a column with a unique identifier (ID) plus a number of variables which are unique to each table. You want to merge them into one big table so that for each ID you have all available variables. You want to make sure that the same ID number (e.g. 108) corresponds to the same case in both datasets, but not all IDs needs to be present in both datasets.
Let’s make up some synthetic data:
a <- data.frame(PID = 101:109,
Hospital = c("UCSF", "HUP", "Stanford",
"Stanford", "UCSF", "HUP",
"HUP", "Stanford", "UCSF"),
Age = c(22, 34, 41, 19, 53, 21, 63, 22, 19),
Sex = c(1, 1, 0, 1, 0, 0, 1, 0, 0))
a PID Hospital Age Sex
1 101 UCSF 22 1
2 102 HUP 34 1
3 103 Stanford 41 0
4 104 Stanford 19 1
5 105 UCSF 53 0
6 106 HUP 21 0
7 107 HUP 63 1
8 108 Stanford 22 0
9 109 UCSF 19 0dim(a)[1] 9 4b <- data.frame(PID = 106:112,
V1 = c(153, 89, 112, 228, 91, 190, 101),
Department = c("Neurology", "Radiology",
"Emergency", "Cardiology",
"Surgery", "Neurology", "Psychiatry"))
dim(b)[1] 7 3b PID V1 Department
1 106 153 Neurology
2 107 89 Radiology
3 108 112 Emergency
4 109 228 Cardiology
5 110 91 Surgery
6 111 190 Neurology
7 112 101 Psychiatry
27.1 merge()
R’s merge() function is used to perform table joins.
Syntax: merge(x, y, by)
where x and y and the two data.frames to join, and by is the column name of the ID variable used to identify rows. If the two datasets’ ID column has a different name, e.g. “PatientID” in one and “PID” in the other, you can either rename one of them to match the other, or use the following syntax:
merge(x, y, by.x, by.y)
where by.x is the name of the ID column for the x dataset and by.y is the name of the ID column for the y dataset.
If you do not specify by or by.x and by.y arguments, merge() defaults to using the intersection of column names of the two input datasets. From merge()’s documentation: by = intersect(names(x), names(y))
In our example datasets above, this works as expected and identifies “PID” as the common column:
27.2 Inner join
The default arguments of merge() perform an inner join:
ab_inner <- merge(a, b)
# same as
ab_inner <- merge(a, b, by = "PID")
# same as
ab_inner <- merge(a, b, all = FALSE)
ab_inner PID Hospital Age Sex V1 Department
1 106 HUP 21 0 153 Neurology
2 107 HUP 63 1 89 Radiology
3 108 Stanford 22 0 112 Emergency
4 109 UCSF 19 0 228 CardiologyNote that the resulting table only contains cases found in both datasets, i.e. IDs 106 through 109
27.3 Outer join
You can perform an outer join by specifying all = TRUE:
ab_outer <- merge(a, b, all = TRUE)
# same as
ab_outer <- merge(a, b, by = "PID", all = TRUE)
ab_outer PID Hospital Age Sex V1 Department
1 101 UCSF 22 1 NA <NA>
2 102 HUP 34 1 NA <NA>
3 103 Stanford 41 0 NA <NA>
4 104 Stanford 19 1 NA <NA>
5 105 UCSF 53 0 NA <NA>
6 106 HUP 21 0 153 Neurology
7 107 HUP 63 1 89 Radiology
8 108 Stanford 22 0 112 Emergency
9 109 UCSF 19 0 228 Cardiology
10 110 <NA> NA NA 91 Surgery
11 111 <NA> NA NA 190 Neurology
12 112 <NA> NA NA 101 PsychiatryNote that the resulting data frame contains all cases found in either dataset and missing values are represented with NA.
27.4 Left outer join
You can perform a left outer join by specifying all.x = TRUE:
ab_leftOuter <- merge(a, b, all.x = TRUE)
ab_leftOuter PID Hospital Age Sex V1 Department
1 101 UCSF 22 1 NA <NA>
2 102 HUP 34 1 NA <NA>
3 103 Stanford 41 0 NA <NA>
4 104 Stanford 19 1 NA <NA>
5 105 UCSF 53 0 NA <NA>
6 106 HUP 21 0 153 Neurology
7 107 HUP 63 1 89 Radiology
8 108 Stanford 22 0 112 Emergency
9 109 UCSF 19 0 228 CardiologyNote that the resulting data frame contains all cases present in the left input dataset (i.e. the one defined first in the arguments) only.
27.5 Right outer join
You can perform a right outer join by specifying all.y = TRUE:
ab_rightOuter <- merge(a, b, all.y = TRUE)
ab_rightOuter PID Hospital Age Sex V1 Department
1 106 HUP 21 0 153 Neurology
2 107 HUP 63 1 89 Radiology
3 108 Stanford 22 0 112 Emergency
4 109 UCSF 19 0 228 Cardiology
5 110 <NA> NA NA 91 Surgery
6 111 <NA> NA NA 190 Neurology
7 112 <NA> NA NA 101 PsychiatryNote how the resulting data frame contains all cases present in the right input dataset (i.e. the one defined second in the arguments) only.
27.6 Specifying columns
As mentioned above, if the ID columns in the two data.frames to be merged do not have the same name, you can specify them directly:
a <- data.frame(PID = 101:109,
Hospital = c("UCSF", "HUP", "Stanford",
"Stanford", "UCSF", "HUP",
"HUP", "Stanford", "UCSF"),
Age = c(22, 34, 41, 19, 53, 21, 63, 22, 19),
Sex = c(1, 1, 0, 1, 0, 0, 1, 0, 0))
a PID Hospital Age Sex
1 101 UCSF 22 1
2 102 HUP 34 1
3 103 Stanford 41 0
4 104 Stanford 19 1
5 105 UCSF 53 0
6 106 HUP 21 0
7 107 HUP 63 1
8 108 Stanford 22 0
9 109 UCSF 19 0b <- data.frame(PatientID = 106:112,
V1 = c(153, 89, 112, 228, 91, 190, 101),
Department = c("Neurology", "Radiology",
"Emergency", "Cardiology",
"Surgery", "Neurology", "Psychiatry"))
b PatientID V1 Department
1 106 153 Neurology
2 107 89 Radiology
3 108 112 Emergency
4 109 228 Cardiology
5 110 91 Surgery
6 111 190 Neurology
7 112 101 Psychiatryab_inner <- merge(a, b, by.x = "PID", by.y = "PatientID")
ab_inner PID Hospital Age Sex V1 Department
1 106 HUP 21 0 153 Neurology
2 107 HUP 63 1 89 Radiology
3 108 Stanford 22 0 112 Emergency
4 109 UCSF 19 0 228 Cardiology27.7 Subsetting
Remember that whatever operation you are performing on one or multiple data.frames, you can always subset rows and/or columns as needed. If, for example, you don’t need to include the “V1” variables in your join, you can directly exclude it:
merge(a, b[, -2], by.x = "PID", by.y = "PatientID") PID Hospital Age Sex Department
1 106 HUP 21 0 Neurology
2 107 HUP 63 1 Radiology
3 108 Stanford 22 0 Emergency
4 109 UCSF 19 0 Cardiology27.8 Joining wide and long tables
The columns defined using the by or by.x and by.y arguments determine which rows from each table to include in the merge. These do not have to identify unique rows in either dataset: for example, if you are merging on a PID column, either table can include repeated PIDs. This allows merging wide and long tables.
As an example, we create a long and a wide table and merge them:
dat_long <- data.frame(
Account_ID = c(8001, 8002, 8003, 8004, 8001, 8002, 8003, 8004,
8001, 8002, 8003, 8004, 8001, 8002, 8003, 8004),
Age = c(67.8017038366664, 42.9198507293701, 46.2301756642422,
39.665983196671, 67.8017038366664, 42.9198507293701,
46.2301756642422, 39.665983196671, 67.8017038366664,
42.9198507293701, 46.2301756642422, 39.665983196671,
67.8017038366664, 42.9198507293701, 46.2301756642422,
39.665983196671),
Admission = c("ED", "Planned", "Planned", "ED", "ED", "Planned",
"Planned", "ED", "ED", "Planned", "Planned", "ED", "ED", "Planned",
"Planned", "ED"),
Lab_key = c("RBC", "RBC", "RBC", "RBC", "WBC", "WBC", "WBC", "WBC",
"Hematocrit", "Hematocrit", "Hematocrit", "Hematocrit",
"Hemoglobin", "Hemoglobin", "Hemoglobin", "Hemoglobin"),
Lab_value = c(4.63449321082268, 3.34968550627897, 4.27037213597765,
4.93897736897793, 8374.22887757195, 7612.37380499927,
8759.27855519425, 6972.28096216548, 36.272693147236,
40.5716317809522, 39.9888624177955, 39.8786884058422,
12.6188444991545, 12.1739747363806, 15.1293426442183,
14.8885696185238)
)
dat_long <- dat_long[order(dat_long$Account_ID), ]
dat_long Account_ID Age Admission Lab_key Lab_value
1 8001 67.80170 ED RBC 4.634493
5 8001 67.80170 ED WBC 8374.228878
9 8001 67.80170 ED Hematocrit 36.272693
13 8001 67.80170 ED Hemoglobin 12.618844
2 8002 42.91985 Planned RBC 3.349686
6 8002 42.91985 Planned WBC 7612.373805
10 8002 42.91985 Planned Hematocrit 40.571632
14 8002 42.91985 Planned Hemoglobin 12.173975
3 8003 46.23018 Planned RBC 4.270372
7 8003 46.23018 Planned WBC 8759.278555
11 8003 46.23018 Planned Hematocrit 39.988862
15 8003 46.23018 Planned Hemoglobin 15.129343
4 8004 39.66598 ED RBC 4.938977
8 8004 39.66598 ED WBC 6972.280962
12 8004 39.66598 ED Hematocrit 39.878688
16 8004 39.66598 ED Hemoglobin 14.888570dat_wide <- data.frame(
Account_ID = c(8002, 8003, 8005),
Department = c("Cardiology", "Neurology", "Surgery"),
Site = c("ZSFG", "Mission_Bay", "Mt_Zion")
)
dat_wide Account_ID Department Site
1 8002 Cardiology ZSFG
2 8003 Neurology Mission_Bay
3 8005 Surgery Mt_ZionInner join:
merge(dat_wide, dat_long) Account_ID Department Site Age Admission Lab_key Lab_value
1 8002 Cardiology ZSFG 42.91985 Planned RBC 3.349686
2 8002 Cardiology ZSFG 42.91985 Planned WBC 7612.373805
3 8002 Cardiology ZSFG 42.91985 Planned Hematocrit 40.571632
4 8002 Cardiology ZSFG 42.91985 Planned Hemoglobin 12.173975
5 8003 Neurology Mission_Bay 46.23018 Planned RBC 4.270372
6 8003 Neurology Mission_Bay 46.23018 Planned WBC 8759.278555
7 8003 Neurology Mission_Bay 46.23018 Planned Hematocrit 39.988862
8 8003 Neurology Mission_Bay 46.23018 Planned Hemoglobin 15.129343Outer join:
merge(dat_wide, dat_long, all = TRUE) Account_ID Department Site Age Admission Lab_key Lab_value
1 8001 <NA> <NA> 67.80170 ED RBC 4.634493
2 8001 <NA> <NA> 67.80170 ED WBC 8374.228878
3 8001 <NA> <NA> 67.80170 ED Hematocrit 36.272693
4 8001 <NA> <NA> 67.80170 ED Hemoglobin 12.618844
5 8002 Cardiology ZSFG 42.91985 Planned RBC 3.349686
6 8002 Cardiology ZSFG 42.91985 Planned WBC 7612.373805
7 8002 Cardiology ZSFG 42.91985 Planned Hematocrit 40.571632
8 8002 Cardiology ZSFG 42.91985 Planned Hemoglobin 12.173975
9 8003 Neurology Mission_Bay 46.23018 Planned RBC 4.270372
10 8003 Neurology Mission_Bay 46.23018 Planned WBC 8759.278555
11 8003 Neurology Mission_Bay 46.23018 Planned Hematocrit 39.988862
12 8003 Neurology Mission_Bay 46.23018 Planned Hemoglobin 15.129343
13 8004 <NA> <NA> 39.66598 ED RBC 4.938977
14 8004 <NA> <NA> 39.66598 ED WBC 6972.280962
15 8004 <NA> <NA> 39.66598 ED Hematocrit 39.878688
16 8004 <NA> <NA> 39.66598 ED Hemoglobin 14.888570
17 8005 Surgery Mt_Zion NA <NA> <NA> NAAs you see above, the output of a join between a wide and long table will be a long table. Depending on the specific analysis goals, one can keep the data in long format, reshape one table prior to merging, or reshape the merged table after joining (See Chapter 26 for reshaping).
27.9 Renaming non-unique columns
Two tables may contain shared column names on columns other than those used for merging. In such cases, the suffixes argument, which defaults to c(".x",".y"), defines the suffix to be added to the left and right tables, respectively.
visit1 <- data.frame(ID = c(8001, 8002, 8003),
Height = c(1.67, 1.79, 1.74),
SBP = c(124, 138, 129))
visit2 <- data.frame(ID = c(8002, 8003, 8004),
Department = c("Cardiology", "Neurology", "Surgery"),
SBP = c(128, 136, 131))
merge(visit1, visit2, by = "ID", suffixes = c("_visit1", "_visit2")) ID Height SBP_visit1 Department SBP_visit2
1 8002 1.79 138 Cardiology 128
2 8003 1.74 129 Neurology 136Note that in these cases, the by argument, or the by.x and by.y arguments, need to be specified so that the other shared column is not erroneously included in the matching.
