26 Reshaping
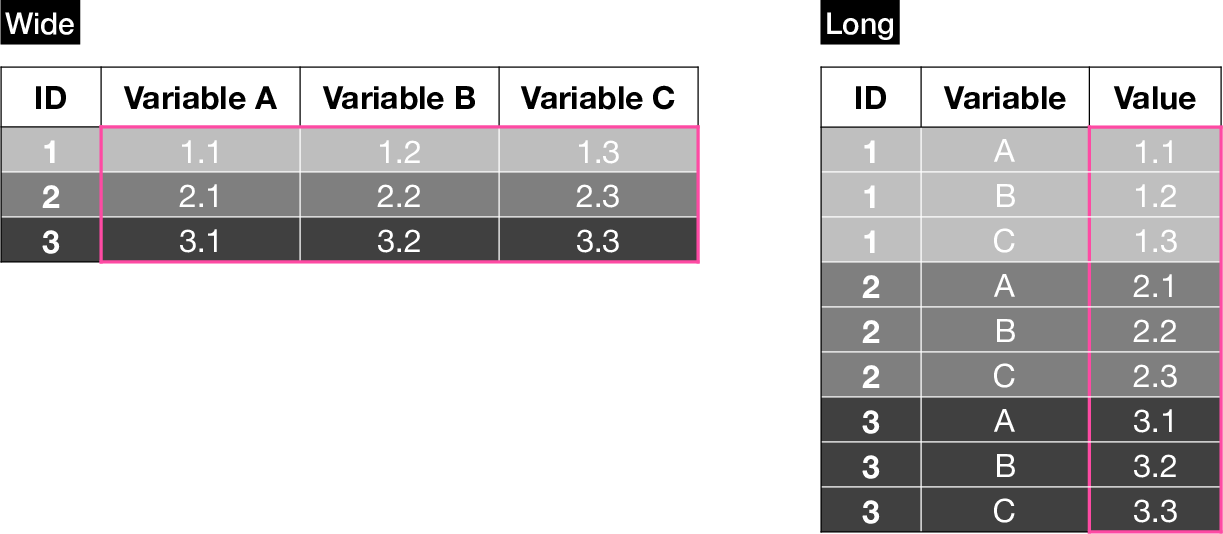
Tabular data can be stored in different formats. Two of the most common ones are wide and long.
A wide dataset contains a single row per case (e.g. patient), while a long dataset can contain multiple rows per case (e.g. for multiple variables or timepoints).
We want to be able to reshape from one form to the other because different programs (e.g. statistical models, plotting functions) expect data in one or the other format for different applications (e.g. longitudinal modeling or grouped visualizations).
R’s reshape() function is very powerful, but can seem intimidating at first, because its documentation is not very clear, especially if you’re not familiar with the jargon.
This chapter includes detailed diagrams and step-by-step instructions to explain how to build calls for long-to-wide and wide-to-long reshaping.
26.1 Long to Wide
26.1.1 Key-value pairs
It is very common to receive data in long format. For example, many tables with electronic health records are stored in long format.
Let’s start with a small synthetic dataset:
dat_long <- data.frame(
Account_ID = c(8001, 8002, 8003, 8004, 8001, 8002, 8003, 8004,
8001, 8002, 8003, 8004, 8001, 8002, 8003, 8004),
Age = c(67.8017038366664, 42.9198507293701, 46.2301756642422,
39.665983196671, 67.8017038366664, 42.9198507293701,
46.2301756642422, 39.665983196671, 67.8017038366664,
42.9198507293701, 46.2301756642422, 39.665983196671,
67.8017038366664, 42.9198507293701, 46.2301756642422,
39.665983196671),
Admission = c("ED", "Planned", "Planned", "ED", "ED", "Planned",
"Planned", "ED", "ED", "Planned", "Planned", "ED", "ED", "Planned",
"Planned", "ED"),
Lab_key = c("RBC", "RBC", "RBC", "RBC", "WBC", "WBC", "WBC", "WBC",
"Hematocrit", "Hematocrit", "Hematocrit", "Hematocrit",
"Hemoglobin", "Hemoglobin", "Hemoglobin", "Hemoglobin"),
Lab_value = c(4.63449321082268, 3.34968550627897, 4.27037213597765,
4.93897736897793, 8374.22887757195, 7612.37380499927,
8759.27855519425, 6972.28096216548, 36.272693147236,
40.5716317809522, 39.9888624177955, 39.8786884058422,
12.6188444991545, 12.1739747363806, 15.1293426442183,
14.8885696185238)
)
dat_long <- dat_long[order(dat_long$Account_ID), ]
dat_long Account_ID Age Admission Lab_key Lab_value
1 8001 67.80170 ED RBC 4.634493
5 8001 67.80170 ED WBC 8374.228878
9 8001 67.80170 ED Hematocrit 36.272693
13 8001 67.80170 ED Hemoglobin 12.618844
2 8002 42.91985 Planned RBC 3.349686
6 8002 42.91985 Planned WBC 7612.373805
10 8002 42.91985 Planned Hematocrit 40.571632
14 8002 42.91985 Planned Hemoglobin 12.173975
3 8003 46.23018 Planned RBC 4.270372
7 8003 46.23018 Planned WBC 8759.278555
11 8003 46.23018 Planned Hematocrit 39.988862
15 8003 46.23018 Planned Hemoglobin 15.129343
4 8004 39.66598 ED RBC 4.938977
8 8004 39.66598 ED WBC 6972.280962
12 8004 39.66598 ED Hematocrit 39.878688
16 8004 39.66598 ED Hemoglobin 14.888570The dataset consists of an Account_ID, denoting a unique patient identifier, Age, Admission, and a pair of Lab_key and a Lab_value columns. The lab data contains information on four lab results: RBC, WBC, Hematocrit, and Hemoglobin.
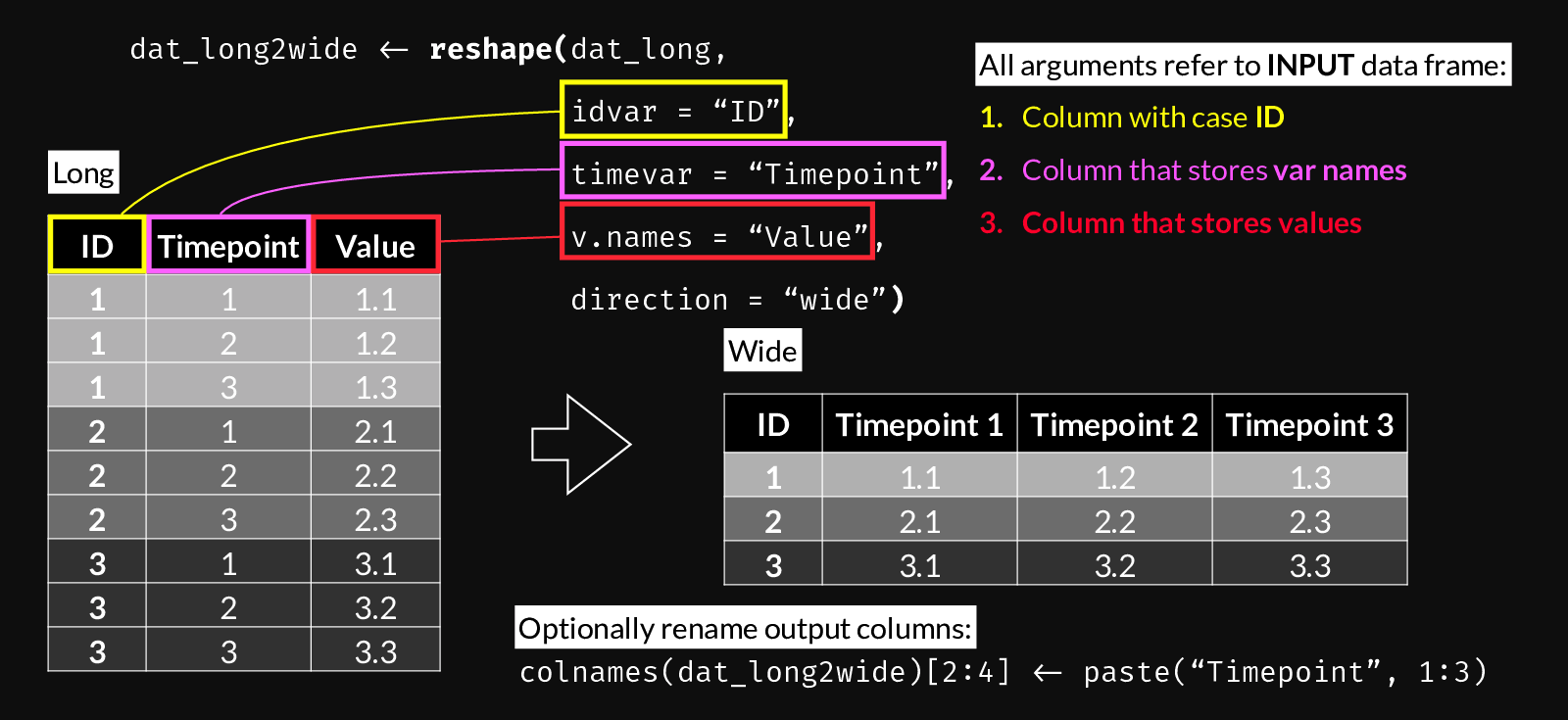
Use the following figure to understand reshape()’s long-to-wide syntax. You can use it as a reference when building a call to reshape() by following steps 1 through 3.
reshape() syntax for Long to Wide transformation.
dat_wide <- reshape(
dat_long,
idvar = "Account_ID",
timevar = "Lab_key",
v.names = "Lab_value",
direction = "wide")
dat_wide Account_ID Age Admission Lab_value.RBC Lab_value.WBC
1 8001 67.80170 ED 4.634493 8374.229
2 8002 42.91985 Planned 3.349686 7612.374
3 8003 46.23018 Planned 4.270372 8759.279
4 8004 39.66598 ED 4.938977 6972.281
Lab_value.Hematocrit Lab_value.Hemoglobin
1 36.27269 12.61884
2 40.57163 12.17397
3 39.98886 15.12934
4 39.87869 14.88857You can optionally clean up column names using gsub(), e.g.
Account_ID Age Admission RBC WBC Hematocrit Hemoglobin
1 8001 67.80170 ED 4.634493 8374.229 36.27269 12.61884
2 8002 42.91985 Planned 3.349686 7612.374 40.57163 12.17397
3 8003 46.23018 Planned 4.270372 8759.279 39.98886 15.12934
4 8004 39.66598 ED 4.938977 6972.281 39.87869 14.8885726.1.2 Incomplete data
It is very common that not all cases have entries for all variables. We can simulate this by removing a few lines from the data frame above.
dat_long <- dat_long[-4, ]
dat_long <- dat_long[-6, ]
dat_long <- dat_long[-13, ]
dat_long Account_ID Age Admission Lab_key Lab_value
1 8001 67.80170 ED RBC 4.634493
5 8001 67.80170 ED WBC 8374.228878
9 8001 67.80170 ED Hematocrit 36.272693
2 8002 42.91985 Planned RBC 3.349686
6 8002 42.91985 Planned WBC 7612.373805
14 8002 42.91985 Planned Hemoglobin 12.173975
3 8003 46.23018 Planned RBC 4.270372
7 8003 46.23018 Planned WBC 8759.278555
11 8003 46.23018 Planned Hematocrit 39.988862
15 8003 46.23018 Planned Hemoglobin 15.129343
4 8004 39.66598 ED RBC 4.938977
8 8004 39.66598 ED WBC 6972.280962
16 8004 39.66598 ED Hemoglobin 14.888570In such cases, long to wide conversion will include NA values where no data is available:
dat_wide <- reshape(dat_long,
idvar = "Account_ID",
timevar = "Lab_key",
v.names = "Lab_value",
direction = "wide"
)
dat_wide Account_ID Age Admission Lab_value.RBC Lab_value.WBC
1 8001 67.80170 ED 4.634493 8374.229
2 8002 42.91985 Planned 3.349686 7612.374
3 8003 46.23018 Planned 4.270372 8759.279
4 8004 39.66598 ED 4.938977 6972.281
Lab_value.Hematocrit Lab_value.Hemoglobin
1 36.27269 NA
2 NA 12.17397
3 39.98886 15.12934
4 NA 14.8885726.1.3 Longitudinal data
dat2 <- data.frame(
pat_enc_csn_id = rep(c(14568:14571), each = 5),
result_date = rep(c(
seq(as.Date("2019-08-01"),
as.Date("2019-08-05"),
length.out = 5
)), 4
),
order_description = rep("WBC", 20),
result_component_value = c(
rnorm(5, mean = 6800, sd = 3840),
rnorm(5, mean = 7900, sd = 3100),
rnorm(5, mean = 8100, sd = 4030),
rnorm(5, mean = 3200, sd = 1100)
))
dat2 pat_enc_csn_id result_date order_description result_component_value
1 14568 2019-08-01 WBC 7772.795
2 14568 2019-08-02 WBC 7146.819
3 14568 2019-08-03 WBC 2529.658
4 14568 2019-08-04 WBC 9592.025
5 14568 2019-08-05 WBC 8195.162
6 14569 2019-08-01 WBC 11049.717
7 14569 2019-08-02 WBC 10922.226
8 14569 2019-08-03 WBC 7416.969
9 14569 2019-08-04 WBC 7024.867
10 14569 2019-08-05 WBC 10343.027
11 14570 2019-08-01 WBC 11415.288
12 14570 2019-08-02 WBC 5936.171
13 14570 2019-08-03 WBC 9115.976
14 14570 2019-08-04 WBC 8360.018
15 14570 2019-08-05 WBC 10205.501
16 14571 2019-08-01 WBC 4030.794
17 14571 2019-08-02 WBC 3444.678
18 14571 2019-08-03 WBC 4044.613
19 14571 2019-08-04 WBC 5882.935
20 14571 2019-08-05 WBC 3439.582In this example, we have four unique patient IDs, with five measurements taken on different days.
Following the same recipe as above, we convert to wide format:
dat2_wide <- reshape(dat2,
idvar = "pat_enc_csn_id",
timevar = "result_date",
v.names = "result_component_value",
direction = "wide"
)
dat2_wide pat_enc_csn_id order_description result_component_value.2019-08-01
1 14568 WBC 7772.795
6 14569 WBC 11049.717
11 14570 WBC 11415.288
16 14571 WBC 4030.794
result_component_value.2019-08-02 result_component_value.2019-08-03
1 7146.819 2529.658
6 10922.226 7416.969
11 5936.171 9115.976
16 3444.678 4044.613
result_component_value.2019-08-04 result_component_value.2019-08-05
1 9592.025 8195.162
6 7024.867 10343.027
11 8360.018 10205.501
16 5882.935 3439.58226.2 Wide to Long
Synthetic data:
set.seed(2022)
dat_wide <- data.frame(
Account_ID = c(8001, 8002, 8003, 8004),
Age = rnorm(4, mean = 57, sd = 12),
RBC = rnorm(4, mean = 4.8, sd = 0.5),
WBC = rnorm(4, mean = 7250, sd = 1500),
Hematocrit = rnorm(4, mean = 40.2, sd = 4),
Hemoglobin = rnorm(4, mean = 13.6, sd = 1.5),
Admission = sample(c("ED", "Planned"), size = 4, replace = TRUE)
)
dat_wide Account_ID Age RBC WBC Hematocrit Hemoglobin Admission
1 8001 67.80170 4.634493 8374.229 36.27269 12.61884 ED
2 8002 42.91985 3.349686 7612.374 40.57163 12.17397 Planned
3 8003 46.23018 4.270372 8759.279 39.98886 15.12934 Planned
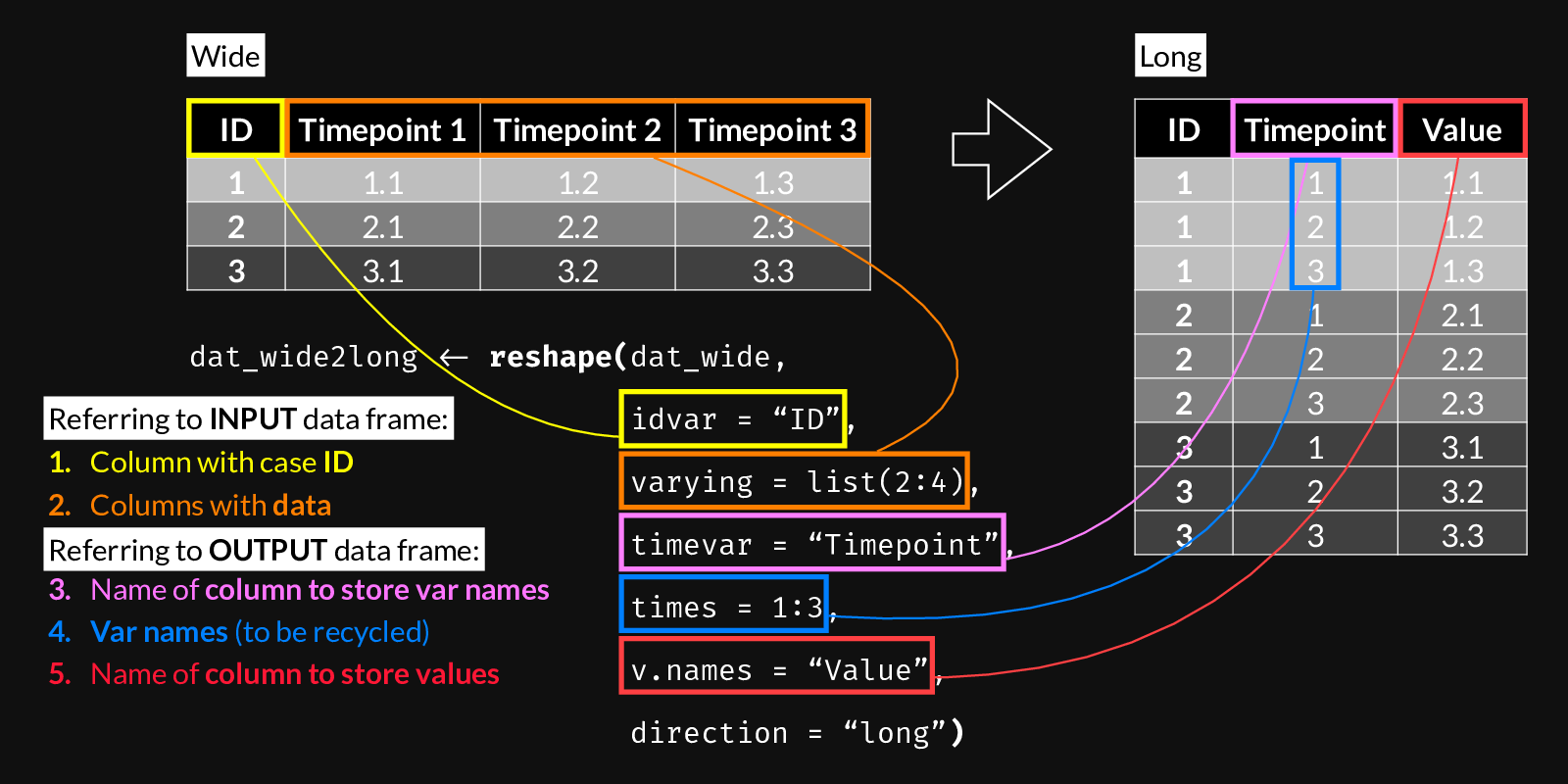
4 8004 39.66598 4.938977 6972.281 39.87869 14.88857 PlannedUse the following figure to understand reshape()’s wide-to-long syntax. Use it as a reference when building a call to reshape() by following steps 1 through 5. It’s important to note which arguments refer to the input vs. the output data frames.
reshape() syntax for Wide to Long transformation.
dat_wide2long <- reshape(# Data in wide format
data = dat_wide,
# The column name that defines case IDs
idvar = "Account_ID",
# The columns whose values we want to keep
varying = list(3:6),
# The name of the new column which will contain all
# the values from the columns above
v.names = "Lab value",
# The values/names, of length = (N columns in "varying"),
# that will be recycled to indicate which column from the
# wide dataset each row corresponds to
times = c(colnames(dat_wide)[3:6]),
# The name of the new column created to hold the values
# defined by "times"
timevar = "Lab key",
direction = "long")
dat_wide2long Account_ID Age Admission Lab key Lab value
8001.RBC 8001 67.80170 ED RBC 4.634493
8002.RBC 8002 42.91985 Planned RBC 3.349686
8003.RBC 8003 46.23018 Planned RBC 4.270372
8004.RBC 8004 39.66598 Planned RBC 4.938977
8001.WBC 8001 67.80170 ED WBC 8374.228878
8002.WBC 8002 42.91985 Planned WBC 7612.373805
8003.WBC 8003 46.23018 Planned WBC 8759.278555
8004.WBC 8004 39.66598 Planned WBC 6972.280962
8001.Hematocrit 8001 67.80170 ED Hematocrit 36.272693
8002.Hematocrit 8002 42.91985 Planned Hematocrit 40.571632
8003.Hematocrit 8003 46.23018 Planned Hematocrit 39.988862
8004.Hematocrit 8004 39.66598 Planned Hematocrit 39.878688
8001.Hemoglobin 8001 67.80170 ED Hemoglobin 12.618844
8002.Hemoglobin 8002 42.91985 Planned Hemoglobin 12.173975
8003.Hemoglobin 8003 46.23018 Planned Hemoglobin 15.129343
8004.Hemoglobin 8004 39.66598 Planned Hemoglobin 14.888570You can also define varying with a character vector:
varying = list(c("RBC", "WBC", "Hematocrit", "Hemoglobin")
Explore the resulting data frame’s attributes:
attributes(dat_wide2long)$row.names
[1] "8001.RBC" "8002.RBC" "8003.RBC" "8004.RBC"
[5] "8001.WBC" "8002.WBC" "8003.WBC" "8004.WBC"
[9] "8001.Hematocrit" "8002.Hematocrit" "8003.Hematocrit" "8004.Hematocrit"
[13] "8001.Hemoglobin" "8002.Hemoglobin" "8003.Hemoglobin" "8004.Hemoglobin"
$names
[1] "Account_ID" "Age" "Admission" "Lab key" "Lab value"
$class
[1] "data.frame"
$reshapeLong
$reshapeLong$varying
$reshapeLong$varying[[1]]
[1] "RBC" "WBC" "Hematocrit" "Hemoglobin"
$reshapeLong$v.names
[1] "Lab value"
$reshapeLong$idvar
[1] "Account_ID"
$reshapeLong$timevar
[1] "Lab key"These attributes are present if and only if a long data.frame was created from a wide data.frame as above. In this case, reshaping back to wide format is as easy as calling reshape() on the previously converted data.frame with no arguments:
dat_wideagain <- reshape(dat_wide2long)
dat_wideagain Account_ID Age Admission RBC WBC Hematocrit Hemoglobin
8001.RBC 8001 67.80170 ED 4.634493 8374.229 36.27269 12.61884
8002.RBC 8002 42.91985 Planned 3.349686 7612.374 40.57163 12.17397
8003.RBC 8003 46.23018 Planned 4.270372 8759.279 39.98886 15.12934
8004.RBC 8004 39.66598 Planned 4.938977 6972.281 39.87869 14.88857Note that the reverse does not work, you need to specify the wide to long reshaping normally.