library(rtemis)18 Additive Tree
The Additive Tree walks like CART, but learns like Gradient Boosting. In other words, it is an algorithm that builds a single decision tree, similar to CART, but the training is similar to boosting stumps (a stump is a tree of depth 1). This results in increased accuracy without sacrificing interpretability (Luna et al. 2019).
As with all supervised learning functions in rtemis, you can either provide a feature matrix / data frame, x, and an outcome vector, y, separately, or provide a combined dataset x alone, in which case the last column should be the outcome.
For classification, the outcome should be a factor where the first level is the ‘positive’ case.
18.1 Train AddTree
Let’s load a dataset from the UCI ML repository:
- We convert the outcome variable “status” to a factor,
- move it to the last column,
- and set levels appropriately
- We then use
check_data()examine the dataset
parkinsons <- read.csv("https://archive.ics.uci.edu/ml/machine-learning-databases/parkinsons/parkinsons.data")
parkinsons$Status <- factor(parkinsons$status, levels = c(1, 0))
parkinsons$status <- NULL
parkinsons$name <- NULL
check_data(parkinsons) parkinsons: A data.table with 195 rows and 23 columns
Data types
* 22 numeric features
* 0 integer features
* 1 factor, which is not ordered
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 0 missing values
Recommendations
* Everything looks good
Let’s train an Additive Tree model on the full sample:
parkinsons.addtree <- s_AddTree(parkinsons,
gamma = .8,
learning.rate = .1)01-07-24 02:01:14 Hello, egenn [s_AddTree]
01-07-24 02:01:14 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 195 x 22
Training outcome: 195 x 1
Testing features: Not available
Testing outcome: Not available
01-07-24 02:01:14 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 145 1
0 2 47
Overall
Sensitivity 0.9864
Specificity 0.9792
Balanced Accuracy 0.9828
PPV 0.9932
NPV 0.9592
F1 0.9898
Accuracy 0.9846
Positive Class: 1
01-07-24 02:01:15 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:15 Converting paths to rules... [s_AddTree]
01-07-24 02:01:15 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:15 Pruning tree... [s_AddTree]
01-07-24 02:01:15 Completed in 0.01 minutes (Real: 0.70; User: 0.68; System: 0.02) [s_AddTree]
18.1.1 Plot AddTree
AddTree trees are saved as data.tree objects. We can plot them using dplot3_addtree(), which creates html output using graphviz.
The first line shows the rule, followed by the N of samples that match the rule, and lastly by the percent of the above that were outcome positive.
By default, leaf nodes with an estimate of 1 (positive class) are orange, and those with estimate 0 are teal.
You can mouse over nodes, edges, and the plot background for some popup info.
dplot3_addtree(parkinsons.addtree)18.1.2 Print AddTree
We can also explore the tree in the console without plotting:
parkinsons.addtree$mod$addtree.pruned levelName
1 All cases
2 ¦--PPE < 0.1339935
3 °--PPE >= 0.1339935
4 ¦--Shimmer.APQ5 < 0.012745
5 ¦ ¦--MDVP.Fo.Hz. < 117.25
6 ¦ ¦ ¦--Shimmer.APQ3 < 0.008825
7 ¦ ¦ ¦ ¦--MDVP.Fo.Hz. < 110.723
8 ¦ ¦ ¦ °--MDVP.Fo.Hz. >= 110.723
9 ¦ ¦ °--Shimmer.APQ3 >= 0.008825
10 ¦ °--MDVP.Fo.Hz. >= 117.25
11 °--Shimmer.APQ5 >= 0.012745 Any attribute can be printed along the hierarchical tree structure:
print(parkinsons.addtree$mod$addtree.pruned, "Estimate") levelName Estimate
1 All cases 0
2 ¦--PPE < 0.1339935 0
3 °--PPE >= 0.1339935 1
4 ¦--Shimmer.APQ5 < 0.012745 1
5 ¦ ¦--MDVP.Fo.Hz. < 117.25 0
6 ¦ ¦ ¦--Shimmer.APQ3 < 0.008825 0
7 ¦ ¦ ¦ ¦--MDVP.Fo.Hz. < 110.723 1
8 ¦ ¦ ¦ °--MDVP.Fo.Hz. >= 110.723 0
9 ¦ ¦ °--Shimmer.APQ3 >= 0.008825 1
10 ¦ °--MDVP.Fo.Hz. >= 117.25 1
11 °--Shimmer.APQ5 >= 0.012745 118.1.3 Predict
To get predicted values, use the predict() S3 generic with the familiar syntax
predict(mod, newdata). If newdata is not supplied, it returns the training set predictions (which we call the ‘fitted’ values):
predict(parkinsons.addtree) [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 0 0 0 0 0 0 1
[38] 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 1
[75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[112] 1 1 1 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
[149] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0
[186] 0 0 0 0 0 0 1 0 0 0
Levels: 1 018.1.4 Training and testing
- Create resamples of our data
- Visualize them (white is testing, teal is training)
- Split data to train and test sets
res <- resample(parkinsons,
n.resamples = 10,
resampler = "kfold",
verbose = TRUE,
seed = 2019)01-07-24 02:01:16 Input contains more than one columns; will stratify on last [resample]
.:Resampling Parameters
n.resamples: 10
resampler: kfold
stratify.var: y
strat.n.bins: 4
01-07-24 02:01:16 Using max n bins possible = 2 [kfold]
01-07-24 02:01:16 Created 10 independent folds [resample]
mplot3_res(res)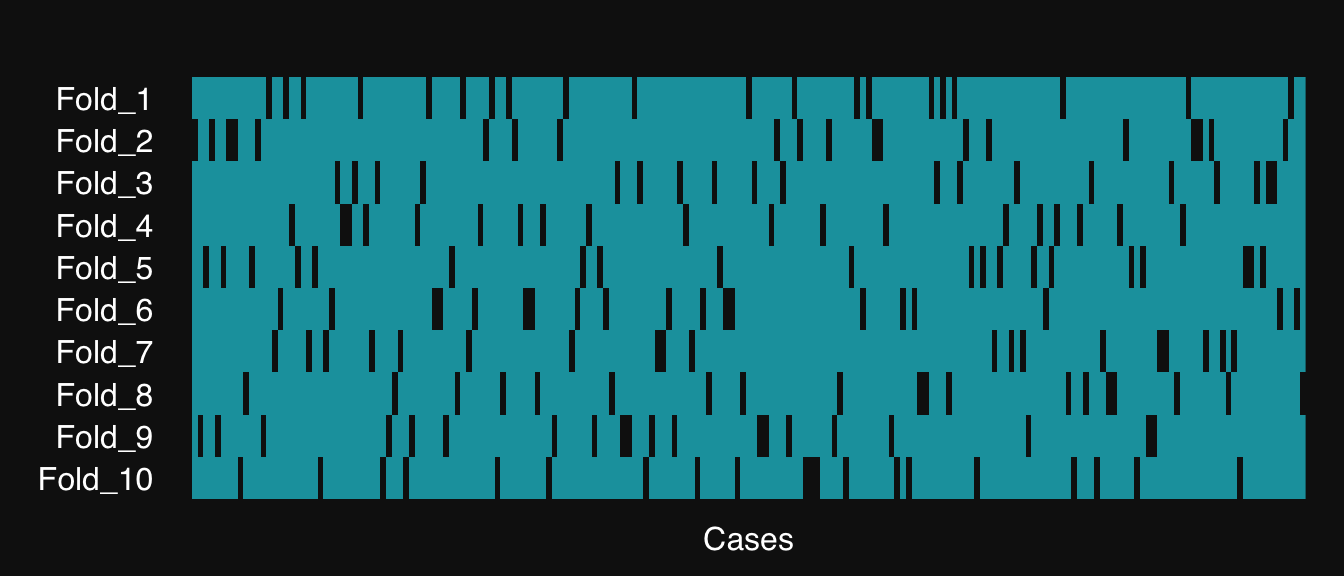
parkinsons.train <- parkinsons[res$Fold_1, ]
parkinsons.test <- parkinsons[-res$Fold_1, ]parkinsons.addtree <- s_AddTree(parkinsons.train, parkinsons.test,
gamma = .8,
learning.rate = .1)01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 20 x 22
Testing outcome: 20 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 130 1
0 2 42
Overall
Sensitivity 0.9848
Specificity 0.9767
Balanced Accuracy 0.9808
PPV 0.9924
NPV 0.9545
F1 0.9886
Accuracy 0.9829
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 13 1
0 2 4
Overall
Sensitivity 0.8667
Specificity 0.8000
Balanced Accuracy 0.8333
PPV 0.9286
NPV 0.6667
F1 0.8966
Accuracy 0.8500
Positive Class: 1
01-07-24 02:01:16 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:16 Converting paths to rules... [s_AddTree]
01-07-24 02:01:16 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:16 Pruning tree... [s_AddTree]
01-07-24 02:01:16 Completed in 4.9e-03 minutes (Real: 0.30; User: 0.29; System: 0.01) [s_AddTree]
18.1.5 Hyperparameter tuning
rtemis supervised learners, like s_AddTree(), support automatic hyperparameter tuning. When more than a single value is passed to a tunable argument, grid search with internal resampling takes place using all available cores (threads).
parkinsons.addtree.tune <- s_AddTree(parkinsons.train, parkinsons.test,
gamma = seq(.6, .9, .1),
learning.rate = .1)01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 20 x 22
Testing outcome: 20 x 1
01-07-24 02:01:16 Running grid search... [gridSearchLearn]
.:Resampling Parameters
n.resamples: 5
resampler: kfold
stratify.var: y
strat.n.bins: 4
01-07-24 02:01:16 Using max n bins possible = 2 [kfold]
01-07-24 02:01:16 Created 5 independent folds [resample]
.:Search parameters
grid.params:
gamma: 0.6, 0.7, 0.8, 0.9
max.depth: 30
learning.rate: 0.1
min.hessian: 0.001
fixed.params:
ifw: TRUE
ifw.type: 2
upsample: FALSE
resample.seed: NULL
01-07-24 02:01:16 Tuning Additive Tree by exhaustive grid search. [gridSearchLearn]
01-07-24 02:01:16 5 inner resamples; 20 models total; running on 8 workers (aarch64-apple-darwin20) [gridSearchLearn]
01-07-24 02:01:16 Running grid line #1 of 20... [...future.FUN]
01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 96 0
0 9 34
Overall
Sensitivity 0.9143
Specificity 1.0000
Balanced Accuracy 0.9571
PPV 1.0000
NPV 0.7907
F1 0.9552
Accuracy 0.9353
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.44; User: 0.35; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #2 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 78 0
0 28 35
Overall
Sensitivity 0.7358
Specificity 1.0000
Balanced Accuracy 0.8679
PPV 1.0000
NPV 0.5556
F1 0.8478
Accuracy 0.8014
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 15 0
0 11 8
Overall
Sensitivity 0.5769
Specificity 1.0000
Balanced Accuracy 0.7885
PPV 1.0000
NPV 0.4211
F1 0.7317
Accuracy 0.6765
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.42; User: 0.39; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #3 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 140 x 22
Training outcome: 140 x 1
Testing features: 35 x 22
Testing outcome: 35 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 102 2
0 4 32
Overall
Sensitivity 0.9623
Specificity 0.9412
Balanced Accuracy 0.9517
PPV 0.9808
NPV 0.8889
F1 0.9714
Accuracy 0.9571
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 0
0 3 9
Overall
Sensitivity 0.8846
Specificity 1.0000
Balanced Accuracy 0.9423
PPV 1.0000
NPV 0.7500
F1 0.9388
Accuracy 0.9143
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:18 Converting paths to rules... [s_AddTree]
01-07-24 02:01:18 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:18 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 0.01 minutes (Real: 0.40; User: 0.39; System: 0.01) [s_AddTree]
01-07-24 02:01:16 Running grid line #4 of 20... [...future.FUN]
01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 96 1
0 10 34
Overall
Sensitivity 0.9057
Specificity 0.9714
Balanced Accuracy 0.9385
PPV 0.9897
NPV 0.7727
F1 0.9458
Accuracy 0.9220
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 3
0 2 5
Overall
Sensitivity 0.9231
Specificity 0.6250
Balanced Accuracy 0.7740
PPV 0.8889
NPV 0.7143
F1 0.9057
Accuracy 0.8529
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.43; User: 0.35; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #5 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 99 2
0 6 32
Overall
Sensitivity 0.9429
Specificity 0.9412
Balanced Accuracy 0.9420
PPV 0.9802
NPV 0.8421
F1 0.9612
Accuracy 0.9424
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 5
0 5 4
Overall
Sensitivity 0.8148
Specificity 0.4444
Balanced Accuracy 0.6296
PPV 0.8148
NPV 0.4444
F1 0.8148
Accuracy 0.7222
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.46; User: 0.43; System: 0.01) [s_AddTree]
01-07-24 02:01:16 Running grid line #6 of 20... [...future.FUN]
01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 97 0
0 8 34
Overall
Sensitivity 0.9238
Specificity 1.0000
Balanced Accuracy 0.9619
PPV 1.0000
NPV 0.8095
F1 0.9604
Accuracy 0.9424
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 4
0 4 5
Overall
Sensitivity 0.8519
Specificity 0.5556
Balanced Accuracy 0.7037
PPV 0.8519
NPV 0.5556
F1 0.8519
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.41; User: 0.33; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #7 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 102 0
0 4 35
Overall
Sensitivity 0.9623
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8974
F1 0.9808
Accuracy 0.9716
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 21 2
0 5 6
Overall
Sensitivity 0.8077
Specificity 0.7500
Balanced Accuracy 0.7788
PPV 0.9130
NPV 0.5455
F1 0.8571
Accuracy 0.7941
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.54; User: 0.51; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #8 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 140 x 22
Training outcome: 140 x 1
Testing features: 35 x 22
Testing outcome: 35 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 102 2
0 4 32
Overall
Sensitivity 0.9623
Specificity 0.9412
Balanced Accuracy 0.9517
PPV 0.9808
NPV 0.8889
F1 0.9714
Accuracy 0.9571
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 0
0 3 9
Overall
Sensitivity 0.8846
Specificity 1.0000
Balanced Accuracy 0.9423
PPV 1.0000
NPV 0.7500
F1 0.9388
Accuracy 0.9143
Positive Class: 1
01-07-24 02:01:18 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:18 Converting paths to rules... [s_AddTree]
01-07-24 02:01:18 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:18 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 0.01 minutes (Real: 0.36; User: 0.35; System: 0.01) [s_AddTree]
01-07-24 02:01:16 Running grid line #9 of 20... [...future.FUN]
01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 99 1
0 7 34
Overall
Sensitivity 0.9340
Specificity 0.9714
Balanced Accuracy 0.9527
PPV 0.9900
NPV 0.8293
F1 0.9612
Accuracy 0.9433
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 2
0 3 6
Overall
Sensitivity 0.8846
Specificity 0.7500
Balanced Accuracy 0.8173
PPV 0.9200
NPV 0.6667
F1 0.9020
Accuracy 0.8529
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.47; User: 0.38; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #10 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 101 2
0 4 32
Overall
Sensitivity 0.9619
Specificity 0.9412
Balanced Accuracy 0.9515
PPV 0.9806
NPV 0.8889
F1 0.9712
Accuracy 0.9568
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 5
0 5 4
Overall
Sensitivity 0.8148
Specificity 0.4444
Balanced Accuracy 0.6296
PPV 0.8148
NPV 0.4444
F1 0.8148
Accuracy 0.7222
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.47; User: 0.44; System: 0.01) [s_AddTree]
01-07-24 02:01:16 Running grid line #11 of 20... [...future.FUN]
01-07-24 02:01:16 Hello, egenn [s_AddTree]
01-07-24 02:01:16 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:16 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 100 0
0 5 34
Overall
Sensitivity 0.9524
Specificity 1.0000
Balanced Accuracy 0.9762
PPV 1.0000
NPV 0.8718
F1 0.9756
Accuracy 0.9640
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.51; User: 0.41; System: 0.08) [s_AddTree]
01-07-24 02:01:17 Running grid line #12 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 102 0
0 4 35
Overall
Sensitivity 0.9623
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8974
F1 0.9808
Accuracy 0.9716
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 2
0 4 6
Overall
Sensitivity 0.8462
Specificity 0.7500
Balanced Accuracy 0.7981
PPV 0.9167
NPV 0.6000
F1 0.8800
Accuracy 0.8235
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 0.01 minutes (Real: 0.50; User: 0.47; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #13 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 140 x 22
Training outcome: 140 x 1
Testing features: 35 x 22
Testing outcome: 35 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 104 1
0 2 33
Overall
Sensitivity 0.9811
Specificity 0.9706
Balanced Accuracy 0.9759
PPV 0.9905
NPV 0.9429
F1 0.9858
Accuracy 0.9786
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 0
0 3 9
Overall
Sensitivity 0.8846
Specificity 1.0000
Balanced Accuracy 0.9423
PPV 1.0000
NPV 0.7500
F1 0.9388
Accuracy 0.9143
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.46; User: 0.37; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #14 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 100 1
0 6 34
Overall
Sensitivity 0.9434
Specificity 0.9714
Balanced Accuracy 0.9574
PPV 0.9901
NPV 0.8500
F1 0.9662
Accuracy 0.9504
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 3
0 2 5
Overall
Sensitivity 0.9231
Specificity 0.6250
Balanced Accuracy 0.7740
PPV 0.8889
NPV 0.7143
F1 0.9057
Accuracy 0.8529
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.34; User: 0.32; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #15 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 103 2
0 2 32
Overall
Sensitivity 0.9810
Specificity 0.9412
Balanced Accuracy 0.9611
PPV 0.9810
NPV 0.9412
F1 0.9810
Accuracy 0.9712
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 2
0 3 7
Overall
Sensitivity 0.8889
Specificity 0.7778
Balanced Accuracy 0.8333
PPV 0.9231
NPV 0.7000
F1 0.9057
Accuracy 0.8611
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:18 Converting paths to rules... [s_AddTree]
01-07-24 02:01:18 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:18 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 0.01 minutes (Real: 0.32; User: 0.31; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #16 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 101 0
0 4 34
Overall
Sensitivity 0.9619
Specificity 1.0000
Balanced Accuracy 0.9810
PPV 1.0000
NPV 0.8947
F1 0.9806
Accuracy 0.9712
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.42; User: 0.34; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #17 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 102 0
0 4 35
Overall
Sensitivity 0.9623
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8974
F1 0.9808
Accuracy 0.9716
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 2
0 4 6
Overall
Sensitivity 0.8462
Specificity 0.7500
Balanced Accuracy 0.7981
PPV 0.9167
NPV 0.6000
F1 0.8800
Accuracy 0.8235
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.45; User: 0.42; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #18 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 140 x 22
Training outcome: 140 x 1
Testing features: 35 x 22
Testing outcome: 35 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 104 0
0 2 34
Overall
Sensitivity 0.9811
Specificity 1.0000
Balanced Accuracy 0.9906
PPV 1.0000
NPV 0.9444
F1 0.9905
Accuracy 0.9857
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 0
0 3 9
Overall
Sensitivity 0.8846
Specificity 1.0000
Balanced Accuracy 0.9423
PPV 1.0000
NPV 0.7500
F1 0.9388
Accuracy 0.9143
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.41; User: 0.34; System: 0.07) [s_AddTree]
01-07-24 02:01:17 Running grid line #19 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 141 x 22
Training outcome: 141 x 1
Testing features: 34 x 22
Testing outcome: 34 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 106 1
0 0 34
Overall
Sensitivity 1.0000
Specificity 0.9714
Balanced Accuracy 0.9857
PPV 0.9907
NPV 1.0000
F1 0.9953
Accuracy 0.9929
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 3
0 2 5
Overall
Sensitivity 0.9231
Specificity 0.6250
Balanced Accuracy 0.7740
PPV 0.8889
NPV 0.7143
F1 0.9057
Accuracy 0.8529
Positive Class: 1
01-07-24 02:01:17 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:17 Converting paths to rules... [s_AddTree]
01-07-24 02:01:17 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:17 Pruning tree... [s_AddTree]
01-07-24 02:01:17 Completed in 0.01 minutes (Real: 0.38; User: 0.36; System: 0.01) [s_AddTree]
01-07-24 02:01:17 Running grid line #20 of 20... [...future.FUN]
01-07-24 02:01:17 Hello, egenn [s_AddTree]
01-07-24 02:01:17 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 139 x 22
Training outcome: 139 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:17 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 103 1
0 2 33
Overall
Sensitivity 0.9810
Specificity 0.9706
Balanced Accuracy 0.9758
PPV 0.9904
NPV 0.9429
F1 0.9856
Accuracy 0.9784
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 2
0 3 7
Overall
Sensitivity 0.8889
Specificity 0.7778
Balanced Accuracy 0.8333
PPV 0.9231
NPV 0.7000
F1 0.9057
Accuracy 0.8611
Positive Class: 1
01-07-24 02:01:18 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:18 Converting paths to rules... [s_AddTree]
01-07-24 02:01:18 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:18 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 4.9e-03 minutes (Real: 0.29; User: 0.28; System: 0.01) [s_AddTree]
.:Best parameters to maximize Balanced Accuracy
best.tune:
gamma: 0.8
max.depth: 30
learning.rate: 0.1
min.hessian: 0.001
01-07-24 02:01:18 Completed in 0.02 minutes (Real: 1.45; User: 0.09; System: 0.11) [gridSearchLearn]
01-07-24 02:01:18 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 130 1
0 2 42
Overall
Sensitivity 0.9848
Specificity 0.9767
Balanced Accuracy 0.9808
PPV 0.9924
NPV 0.9545
F1 0.9886
Accuracy 0.9829
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 13 1
0 2 4
Overall
Sensitivity 0.8667
Specificity 0.8000
Balanced Accuracy 0.8333
PPV 0.9286
NPV 0.6667
F1 0.8966
Accuracy 0.8500
Positive Class: 1
01-07-24 02:01:18 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:18 Converting paths to rules... [s_AddTree]
01-07-24 02:01:18 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:18 Pruning tree... [s_AddTree]
01-07-24 02:01:18 Completed in 0.03 minutes (Real: 1.78; User: 0.39; System: 0.14) [s_AddTree]
We can define tuning resampling parameters with the grid.resampler.rtSet argument. The setup.resample() convenience function helps easily build the list needed by grid.resampler.params, providing auto-completion.
parkinsons.addtree.tune <- s_AddTree(parkinsons.train, parkinsons.test,
gamma = seq(.6, .9, .1),
learning.rate = .1,
grid.resample.params = setup.resample(resampler = 'strat.boot',
n.resamples = 5))01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 20 x 22
Testing outcome: 20 x 1
01-07-24 02:01:19 Running grid search... [gridSearchLearn]
.:Resampling Parameters
n.resamples: 5
resampler: strat.boot
stratify.var: y
train.p: 0.8
strat.n.bins: 4
target.length: 175
01-07-24 02:01:19 Using max n bins possible = 2 [strat.sub]
01-07-24 02:01:19 Created 5 stratified bootstraps [resample]
.:Search parameters
grid.params:
gamma: 0.6, 0.7, 0.8, 0.9
max.depth: 30
learning.rate: 0.1
min.hessian: 0.001
fixed.params:
ifw: TRUE
ifw.type: 2
upsample: FALSE
resample.seed: NULL
01-07-24 02:01:19 Tuning Additive Tree by exhaustive grid search. [gridSearchLearn]
01-07-24 02:01:19 5 inner resamples; 20 models total; running on 8 workers (aarch64-apple-darwin20) [gridSearchLearn]
01-07-24 02:01:19 Running grid line #1 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 125 0
0 7 43
Overall
Sensitivity 0.9470
Specificity 1.0000
Balanced Accuracy 0.9735
PPV 1.0000
NPV 0.8600
F1 0.9728
Accuracy 0.9600
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 21 1
0 6 8
Overall
Sensitivity 0.7778
Specificity 0.8889
Balanced Accuracy 0.8333
PPV 0.9545
NPV 0.5714
F1 0.8571
Accuracy 0.8056
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.53; User: 0.43; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #2 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 126 2
0 8 39
Overall
Sensitivity 0.9403
Specificity 0.9512
Balanced Accuracy 0.9458
PPV 0.9844
NPV 0.8298
F1 0.9618
Accuracy 0.9429
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 4
0 4 5
Overall
Sensitivity 0.8519
Specificity 0.5556
Balanced Accuracy 0.7037
PPV 0.8519
NPV 0.5556
F1 0.8519
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.48; User: 0.46; System: 0.01) [s_AddTree]
01-07-24 02:01:20 Running grid line #3 of 20... [...future.FUN]
01-07-24 02:01:20 Hello, egenn [s_AddTree]
01-07-24 02:01:20 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:20 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 120 1
0 11 43
Overall
Sensitivity 0.9160
Specificity 0.9773
Balanced Accuracy 0.9467
PPV 0.9917
NPV 0.7963
F1 0.9524
Accuracy 0.9314
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 2
0 4 7
Overall
Sensitivity 0.8519
Specificity 0.7778
Balanced Accuracy 0.8148
PPV 0.9200
NPV 0.6364
F1 0.8846
Accuracy 0.8333
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.38; User: 0.37; System: 3e-03) [s_AddTree]
01-07-24 02:01:19 Running grid line #4 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 110 0
0 17 48
Overall
Sensitivity 0.8661
Specificity 1.0000
Balanced Accuracy 0.9331
PPV 1.0000
NPV 0.7385
F1 0.9283
Accuracy 0.9029
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 20 1
0 7 8
Overall
Sensitivity 0.7407
Specificity 0.8889
Balanced Accuracy 0.8148
PPV 0.9524
NPV 0.5333
F1 0.8333
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.47; User: 0.38; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #5 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 123 3
0 8 41
Overall
Sensitivity 0.9389
Specificity 0.9318
Balanced Accuracy 0.9354
PPV 0.9762
NPV 0.8367
F1 0.9572
Accuracy 0.9371
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 21 4
0 6 5
Overall
Sensitivity 0.7778
Specificity 0.5556
Balanced Accuracy 0.6667
PPV 0.8400
NPV 0.4545
F1 0.8077
Accuracy 0.7222
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.40; User: 0.37; System: 0.01) [s_AddTree]
01-07-24 02:01:19 Running grid line #6 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 127 0
0 5 43
Overall
Sensitivity 0.9621
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8958
F1 0.9807
Accuracy 0.9714
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 24 2
0 3 7
Overall
Sensitivity 0.8889
Specificity 0.7778
Balanced Accuracy 0.8333
PPV 0.9231
NPV 0.7000
F1 0.9057
Accuracy 0.8611
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.61; User: 0.52; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #7 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 127 0
0 7 41
Overall
Sensitivity 0.9478
Specificity 1.0000
Balanced Accuracy 0.9739
PPV 1.0000
NPV 0.8542
F1 0.9732
Accuracy 0.9600
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 2
0 4 7
Overall
Sensitivity 0.8519
Specificity 0.7778
Balanced Accuracy 0.8148
PPV 0.9200
NPV 0.6364
F1 0.8846
Accuracy 0.8333
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.46; User: 0.42; System: 0.01) [s_AddTree]
01-07-24 02:01:20 Running grid line #8 of 20... [...future.FUN]
01-07-24 02:01:20 Hello, egenn [s_AddTree]
01-07-24 02:01:20 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:20 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 125 1
0 6 43
Overall
Sensitivity 0.9542
Specificity 0.9773
Balanced Accuracy 0.9657
PPV 0.9921
NPV 0.8776
F1 0.9728
Accuracy 0.9600
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 4.7e-03 minutes (Real: 0.28; User: 0.28; System: 3e-03) [s_AddTree]
01-07-24 02:01:19 Running grid line #9 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 121 0
0 6 48
Overall
Sensitivity 0.9528
Specificity 1.0000
Balanced Accuracy 0.9764
PPV 1.0000
NPV 0.8889
F1 0.9758
Accuracy 0.9657
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 1
0 5 8
Overall
Sensitivity 0.8148
Specificity 0.8889
Balanced Accuracy 0.8519
PPV 0.9565
NPV 0.6154
F1 0.8800
Accuracy 0.8333
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.55; User: 0.46; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #10 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 123 2
0 8 42
Overall
Sensitivity 0.9389
Specificity 0.9545
Balanced Accuracy 0.9467
PPV 0.9840
NPV 0.8400
F1 0.9609
Accuracy 0.9429
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 21 4
0 6 5
Overall
Sensitivity 0.7778
Specificity 0.5556
Balanced Accuracy 0.6667
PPV 0.8400
NPV 0.4545
F1 0.8077
Accuracy 0.7222
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.32; User: 0.31; System: 4e-03) [s_AddTree]
01-07-24 02:01:19 Running grid line #11 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 127 0
0 5 43
Overall
Sensitivity 0.9621
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8958
F1 0.9807
Accuracy 0.9714
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 1
0 4 8
Overall
Sensitivity 0.8519
Specificity 0.8889
Balanced Accuracy 0.8704
PPV 0.9583
NPV 0.6667
F1 0.9020
Accuracy 0.8611
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.51; User: 0.43; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #12 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 133 1
0 1 40
Overall
Sensitivity 0.9925
Specificity 0.9756
Balanced Accuracy 0.9841
PPV 0.9925
NPV 0.9756
F1 0.9925
Accuracy 0.9886
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 23 3
0 4 6
Overall
Sensitivity 0.8519
Specificity 0.6667
Balanced Accuracy 0.7593
PPV 0.8846
NPV 0.6000
F1 0.8679
Accuracy 0.8056
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.52; User: 0.50; System: 0.01) [s_AddTree]
01-07-24 02:01:19 Running grid line #13 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 126 1
0 5 43
Overall
Sensitivity 0.9618
Specificity 0.9773
Balanced Accuracy 0.9696
PPV 0.9921
NPV 0.8958
F1 0.9767
Accuracy 0.9657
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.35; User: 0.27; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #14 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 125 0
0 2 48
Overall
Sensitivity 0.9843
Specificity 1.0000
Balanced Accuracy 0.9921
PPV 1.0000
NPV 0.9600
F1 0.9921
Accuracy 0.9886
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 25 1
0 2 8
Overall
Sensitivity 0.9259
Specificity 0.8889
Balanced Accuracy 0.9074
PPV 0.9615
NPV 0.8000
F1 0.9434
Accuracy 0.9167
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.40; User: 0.39; System: 0.01) [s_AddTree]
01-07-24 02:01:19 Running grid line #15 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 128 2
0 3 42
Overall
Sensitivity 0.9771
Specificity 0.9545
Balanced Accuracy 0.9658
PPV 0.9846
NPV 0.9333
F1 0.9808
Accuracy 0.9714
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 20 4
0 7 5
Overall
Sensitivity 0.7407
Specificity 0.5556
Balanced Accuracy 0.6481
PPV 0.8333
NPV 0.4167
F1 0.7843
Accuracy 0.6944
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.37; User: 0.35; System: 0.01) [s_AddTree]
01-07-24 02:01:19 Running grid line #16 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 127 0
0 5 43
Overall
Sensitivity 0.9621
Specificity 1.0000
Balanced Accuracy 0.9811
PPV 1.0000
NPV 0.8958
F1 0.9807
Accuracy 0.9714
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 0
0 5 9
Overall
Sensitivity 0.8148
Specificity 1.0000
Balanced Accuracy 0.9074
PPV 1.0000
NPV 0.6429
F1 0.8980
Accuracy 0.8611
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.55; User: 0.46; System: 0.07) [s_AddTree]
01-07-24 02:01:19 Running grid line #17 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 133 1
0 1 40
Overall
Sensitivity 0.9925
Specificity 0.9756
Balanced Accuracy 0.9841
PPV 0.9925
NPV 0.9756
F1 0.9925
Accuracy 0.9886
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.46; User: 0.44; System: 0.01) [s_AddTree]
01-07-24 02:01:19 Running grid line #18 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 126 1
0 5 43
Overall
Sensitivity 0.9618
Specificity 0.9773
Balanced Accuracy 0.9696
PPV 0.9921
NPV 0.8958
F1 0.9767
Accuracy 0.9657
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 3
0 5 6
Overall
Sensitivity 0.8148
Specificity 0.6667
Balanced Accuracy 0.7407
PPV 0.8800
NPV 0.5455
F1 0.8462
Accuracy 0.7778
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.31; User: 0.23; System: 0.06) [s_AddTree]
01-07-24 02:01:19 Running grid line #19 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 125 0
0 2 48
Overall
Sensitivity 0.9843
Specificity 1.0000
Balanced Accuracy 0.9921
PPV 1.0000
NPV 0.9600
F1 0.9921
Accuracy 0.9886
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 25 4
0 2 5
Overall
Sensitivity 0.9259
Specificity 0.5556
Balanced Accuracy 0.7407
PPV 0.8621
NPV 0.7143
F1 0.8929
Accuracy 0.8333
Positive Class: 1
01-07-24 02:01:19 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:19 Converting paths to rules... [s_AddTree]
01-07-24 02:01:19 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:19 Pruning tree... [s_AddTree]
01-07-24 02:01:19 Completed in 0.01 minutes (Real: 0.38; User: 0.36; System: 5e-03) [s_AddTree]
01-07-24 02:01:19 Running grid line #20 of 20... [...future.FUN]
01-07-24 02:01:19 Hello, egenn [s_AddTree]
01-07-24 02:01:19 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 175 x 22
Training outcome: 175 x 1
Testing features: 36 x 22
Testing outcome: 36 x 1
01-07-24 02:01:19 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 128 1
0 3 43
Overall
Sensitivity 0.9771
Specificity 0.9773
Balanced Accuracy 0.9772
PPV 0.9922
NPV 0.9348
F1 0.9846
Accuracy 0.9771
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 22 4
0 5 5
Overall
Sensitivity 0.8148
Specificity 0.5556
Balanced Accuracy 0.6852
PPV 0.8462
NPV 0.5000
F1 0.8302
Accuracy 0.7500
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.01 minutes (Real: 0.32; User: 0.31; System: 0.01) [s_AddTree]
.:Best parameters to maximize Balanced Accuracy
best.tune:
gamma: 0.8
max.depth: 30
learning.rate: 0.1
min.hessian: 0.001
01-07-24 02:01:20 Completed in 0.02 minutes (Real: 1.45; User: 0.06; System: 0.13) [gridSearchLearn]
01-07-24 02:01:20 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 130 1
0 2 42
Overall
Sensitivity 0.9848
Specificity 0.9767
Balanced Accuracy 0.9808
PPV 0.9924
NPV 0.9545
F1 0.9886
Accuracy 0.9829
Positive Class: 1
.:AddTree Classification Testing Summary
Reference
Estimated 1 0
1 13 1
0 2 4
Overall
Sensitivity 0.8667
Specificity 0.8000
Balanced Accuracy 0.8333
PPV 0.9286
NPV 0.6667
F1 0.8966
Accuracy 0.8500
Positive Class: 1
01-07-24 02:01:20 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:20 Converting paths to rules... [s_AddTree]
01-07-24 02:01:20 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:20 Pruning tree... [s_AddTree]
01-07-24 02:01:20 Completed in 0.03 minutes (Real: 1.77; User: 0.35; System: 0.15) [s_AddTree]
Let’s look at the tuning results (this is a small dataset and tuning may not be very accurate):
parkinsons.addtree.tune$extra$gridSearch$tune.resultsNULL18.1.6 Nested resampling: Cross-validation and hyperparameter tuning
We now use the core rtemis supervised learning function train to use nested resampling for cross-validation and hyperparameter tuning:
parkinsons.addtree.10fold <- train_cv(parkinsons,
mod = 'addtree',
gamma = c(.8, .9),
learning.rate = c(.01, .05),
seed = 2018)01-07-24 02:01:21 Hello, egenn [train_cv]
.:Classification Input Summary
Training features: 195 x 22
Training outcome: 195 x 1
01-07-24 02:01:21 Training Ranger Random Forest on 10 stratified subsamples... [train_cv]
01-07-24 02:01:21 Outer resampling plan set to sequential [resLearn]
.:Cross-validated Ranger
Mean Balanced Accuracy of 10 stratified subsamples: 0.85
01-07-24 02:01:22 Completed in 0.02 minutes (Real: 1.07; User: 1.58; System: 0.47) [train_cv]
We can get a summary of the cross-validation by printing the train object:
parkinsons.addtree.10fold.:rtemis Cross-Validated Classification Model
Ranger (Ranger Random Forest)
Outer resampling: 10 stratified subsamples (1 repeat)
Mean Balanced Accuracy: 0.8516667
18.2 Bagging the Additive Tree (Addtree Random Forest)
You can use rtemis’ bag function to build a random forest with AddTree base learners.
data(Sonar, package = 'mlbench')
res <- resample(Sonar, seed = 2020)01-07-24 02:01:22 Input contains more than one columns; will stratify on last [resample]
.:Resampling Parameters
n.resamples: 10
resampler: strat.sub
stratify.var: y
train.p: 0.75
strat.n.bins: 4
01-07-24 02:01:22 Using max n bins possible = 2 [strat.sub]
01-07-24 02:01:22 Created 10 stratified subsamples [resample]
sonar.train <- Sonar[res$Subsample_1, ]
sonar.test <- Sonar[-res$Subsample_1, ]mod.addtree <- s_AddTree(sonar.train, sonar.test)01-07-24 02:01:22 Hello, egenn [s_AddTree]
01-07-24 02:01:22 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 155 x 60
Training outcome: 155 x 1
Testing features: 53 x 60
Testing outcome: 53 x 1
01-07-24 02:01:22 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated M R
M 82 2
R 1 70
Overall
Sensitivity 0.9880
Specificity 0.9722
Balanced Accuracy 0.9801
PPV 0.9762
NPV 0.9859
F1 0.9820
Accuracy 0.9806
Positive Class: M
.:AddTree Classification Testing Summary
Reference
Estimated M R
M 19 4
R 9 21
Overall
Sensitivity 0.6786
Specificity 0.8400
Balanced Accuracy 0.7593
PPV 0.8261
NPV 0.7000
F1 0.7451
Accuracy 0.7547
Positive Class: M
01-07-24 02:01:23 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:23 Converting paths to rules... [s_AddTree]
01-07-24 02:01:23 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:23 Pruning tree... [s_AddTree]
01-07-24 02:01:23 Completed in 0.01 minutes (Real: 0.61; User: 0.59; System: 0.01) [s_AddTree]
Let’s train a random forest using AddTree base learner (with just 20 trees for this example)
bag.addtree <- bag(sonar.train,
sonar.test,
mod = "AddTree",
k = 20,
mtry = 5)01-07-24 02:01:24 Hello, egenn [bag]
01-07-24 02:01:24 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 155 x 60
Training outcome: 155 x 1
Testing features: 53 x 60
Testing outcome: 53 x 1
.:Parameters
mod: AddTree
mod.params: (empty list)
01-07-24 02:01:24 Bagging 20 Additive Tree... [bag]
01-07-24 02:01:24 Using max n bins possible = 2 [strat.sub]
01-07-24 02:01:24 Outer resampling: Future plan set to multicore with 8 workers [resLearn]
.:Classification Training Summary
Reference
Estimated M R
M 81 1
R 2 71
Overall
Sensitivity 0.9759
Specificity 0.9861
Balanced Accuracy 0.9810
PPV 0.9878
NPV 0.9726
F1 0.9818
Accuracy 0.9806
Positive Class: M
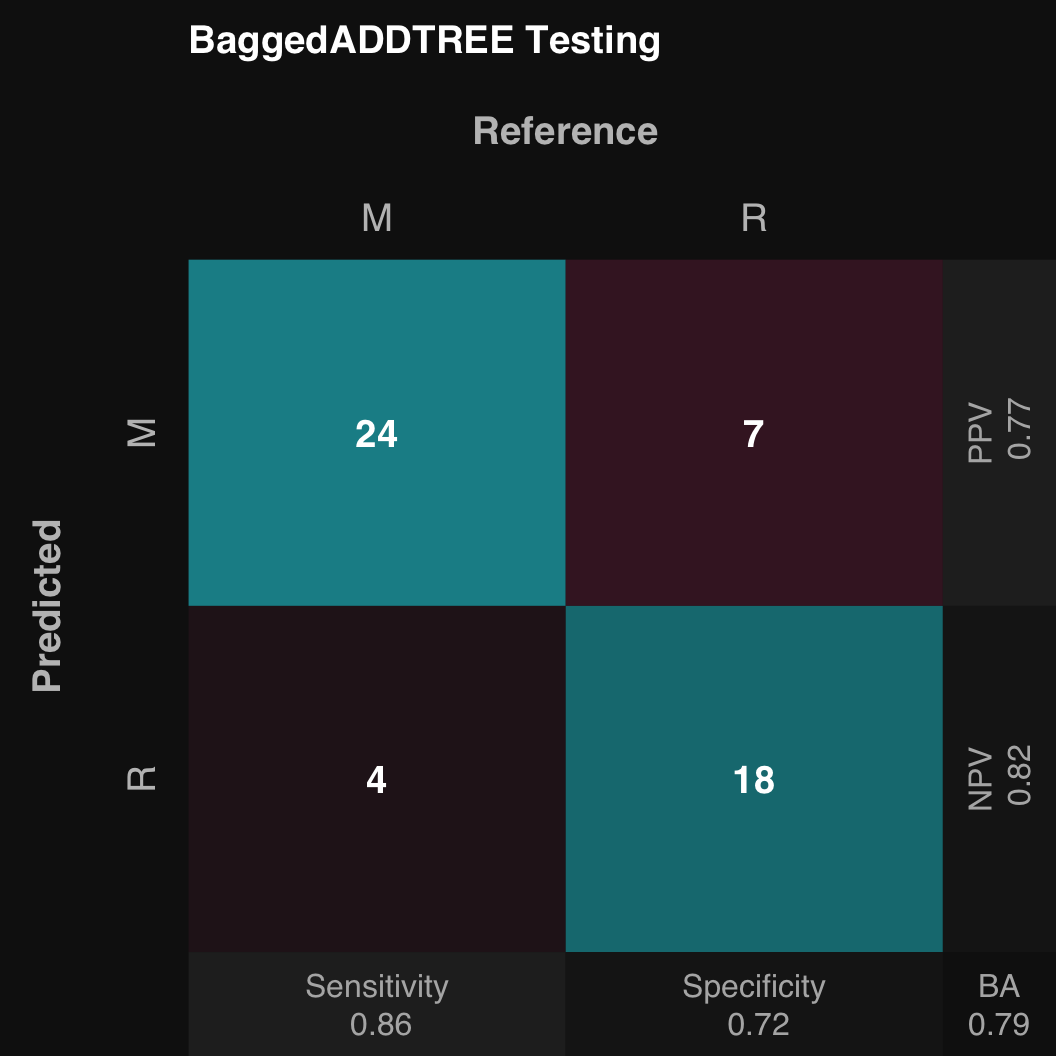
.:Classification Testing Summary
Reference
Estimated M R
M 24 7
R 4 18
Overall
Sensitivity 0.8571
Specificity 0.7200
Balanced Accuracy 0.7886
PPV 0.7742
NPV 0.8182
F1 0.8136
Accuracy 0.7925
Positive Class: M
01-07-24 02:01:27 Completed in 0.06 minutes (Real: 3.62; User: 1.20; System: 0.40) [bag]
18.3 More example datasets
18.3.1 OpenML: sleep
Let’s grab a dataset from the massive OpenML repository.
sleep <- read("https://www.openml.org/data/get_csv/53273/sleep.arff",
na.strings = "?", character2factor = TRUE)01-07-24 02:01:39 ▶ Reading sleep.arff using data.table... [read]
01-07-24 02:01:39 Read in 62 x 8 [read]
01-07-24 02:01:39 Hello, egenn [preprocess]
01-07-24 02:01:39 Converting 1 character feature to a factor... [preprocess]
01-07-24 02:01:39 Completed in 1.7e-05 minutes (Real: 1e-03; User: 2e-03; System: 0.00) [preprocess]
01-07-24 02:01:39 Completed in 3.2e-03 minutes (Real: 0.19; User: 0.01; System: 3e-03) [read]
check_data(sleep) sleep: A data.table with 62 rows and 8 columns
Data types
* 4 numeric features
* 3 integer features
* 1 factor, which is not ordered
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 2 features include 'NA' values; 8 'NA' values total
* 2 numeric
Recommendations
* Consider imputing missing values or use complete cases only
We can impute missing data with preprocess():
sleep <- preprocess(sleep, impute = TRUE)01-07-24 02:01:39 Hello, egenn [preprocess]
01-07-24 02:01:39 Imputing missing values using predictive mean matching with missRanger... [preprocess]
Missing value imputation by random forests
Variables to impute: max_life_span, gestation_time
Variables used to impute: body_weight, brain_weight, max_life_span, gestation_time, predation_index, sleep_exposure_index, danger_index, binaryClass
iter 1
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
iter 2
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
iter 3
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
iter 4
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
01-07-24 02:01:39 Completed in 1.2e-03 minutes (Real: 0.07; User: 0.16; System: 0.08) [preprocess]
Train and plot AddTree:
sleep.addtree <- s_AddTree(sleep, gamma = .8, learning.rate = .1)01-07-24 02:01:39 Hello, egenn [s_AddTree]
01-07-24 02:01:39 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 62 x 7
Training outcome: 62 x 1
Testing features: Not available
Testing outcome: Not available
01-07-24 02:01:39 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated N P
N 31 3
P 2 26
Overall
Sensitivity 0.9394
Specificity 0.8966
Balanced Accuracy 0.9180
PPV 0.9118
NPV 0.9286
F1 0.9254
Accuracy 0.9194
Positive Class: N
01-07-24 02:01:39 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:39 Converting paths to rules... [s_AddTree]
01-07-24 02:01:39 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:39 Pruning tree... [s_AddTree]
01-07-24 02:01:39 Completed in 4.6e-03 minutes (Real: 0.28; User: 0.27; System: 3e-03) [s_AddTree]
dplot3_addtree(sleep.addtree)18.3.2 PMLB: chess
Let’s load a dataset from the Penn ML Benchmarks github repository.
R allows us to read a gzipped file and unzip on the fly:
- We open a remote connection to a gzipped tab-separated file,
- read it in R with
read.table, - set the target levels,
- and check the data
rzd <- gzcon(
url("https://github.com/EpistasisLab/pmlb/raw/master/datasets/chess/chess.tsv.gz"),
text = TRUE)
chess <- read.table(rzd, header = TRUE)
chess$target <- factor(chess$target, levels = c(1, 0))
check_data(chess) chess: A data.table with 3196 rows and 37 columns
Data types
* 0 numeric features
* 36 integer features
* 1 factor, which is not ordered
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 0 missing values
Recommendations
* Everything looks good
chess.addtree <- s_AddTree(chess,
gamma = .8, learning.rate = .1)01-07-24 02:01:40 Hello, egenn [s_AddTree]
01-07-24 02:01:40 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.:Classification Input Summary
Training features: 3196 x 36
Training outcome: 3196 x 1
Testing features: Not available
Testing outcome: Not available
01-07-24 02:01:40 Training AddTree... [s_AddTree]
.:AddTree Classification Training Summary
Reference
Estimated 1 0
1 1623 28
0 46 1499
Overall
Sensitivity 0.9724
Specificity 0.9817
Balanced Accuracy 0.9771
PPV 0.9830
NPV 0.9702
F1 0.9777
Accuracy 0.9768
Positive Class: 1
01-07-24 02:01:43 Traversing tree by preorder... [s_AddTree]
01-07-24 02:01:43 Converting paths to rules... [s_AddTree]
01-07-24 02:01:43 Converting to data.tree object... [s_AddTree]
01-07-24 02:01:43 Pruning tree... [s_AddTree]
01-07-24 02:01:43 Completed in 0.05 minutes (Real: 3.04; User: 2.92; System: 0.09) [s_AddTree]
dplot3_addtree(chess.addtree)