library(rtemis) .:rtemis 0.97.4 🌊 aarch64-apple-darwin20library(rtemis) .:rtemis 0.97.4 🌊 aarch64-apple-darwin20rtemis makes extensive use of R6 classes. (Early in development, methods were created for all available class systems - S3, S4, RC, R6 - and R6 was the winner).
The following classes are defined - you don’t need to learn or remember these, they are created automatically, as appropriate:
rtMod: Supervised model class
rtModClass: Inherits from rtMod and adds support for classification modelsrtModBag: Inherits from rtMod and adds support for bagged modelsrtMeta: Inherits from rtMod and adds support for meta modelsrtModLite: A “lite”, bare-bones version of rtMod, used internally in some applications
rtModCV: Cross-validated models
rtModCVclass: Inherits from rtModCV and adds support for cross-validated classification modelsrtClust: Clustering class
rtDecom: Decomposition class
rtXDecom: Cross-decomposition class
rtMeta: Meta model class
One of the advantages of such a class system is that it allows storing both attributes (e.g. data like fitted values) and methods (functions that can be performed on the object, like plotting) in an object. Regular R methods (like predict, summary, etc), known as S3 generics, are fully compatible with the R6 system.
Let’s look at an example object.
x <- rnormmat(200, 5)
w <- rnorm(5)
y <- x %*% w + rnorm(200)
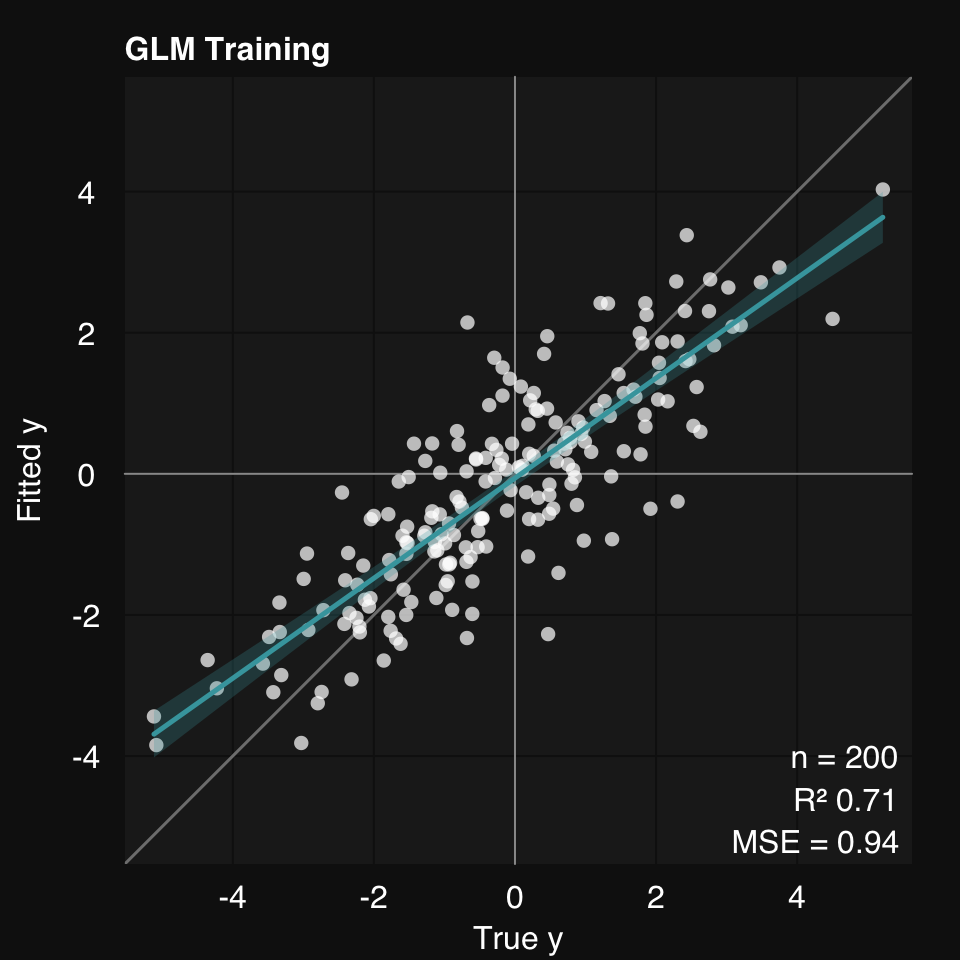
mod <- s_GLM(x, y)06-30-24 10:57:53 Hello, egenn [s_GLM]
.:Regression Input Summary
Training features: 200 x 5
Training outcome: 200 x 1
Testing features: Not available
Testing outcome: Not available
06-30-24 10:57:53 Training GLM... [s_GLM]
.:GLM Regression Training Summary
MSE = 0.94 (70.92%)
RMSE = 0.97 (46.07%)
MAE = 0.77 (46.20%)
r = 0.84 (p = 5.3e-55)
R sq = 0.71
06-30-24 10:57:53 Completed in 0.01 minutes (Real: 0.45; User: 0.40; System: 0.02) [s_GLM]
class(mod)[1] "rtMod" "R6" Let’s look at some of the object attributes Remember, in rtemis, fitted refers to the estimated values for the training set and predicted referes to the estimated values for the test set.
head(mod$fitted)[1] 0.7002195 1.3587610 0.3109817 -1.1798197 1.8645119 0.6057615mod$error.train MSE = 0.94 (70.92%)
RMSE = 0.97 (46.07%)
MAE = 0.77 (46.20%)
r = 0.84 (p = 5.3e-55)
R sq = 0.71
By the way - you notice the error was custom printed.
class(mod$error.train)[1] "regError" "data.frame"It is a simple S3 object of class regError to allow this pretty-printing. You can view the data.frame itself too. In this case, it holds some more information.
as.data.frame(mod$error.train) MAE MSE RMSE NRMSE MAE.EXP MAE.RED MSE.EXP
1 0.7724695 0.9413956 0.9702554 0.09390814 1.435863 0.4620173 3.236854
MSE.RED RMSE.EXP RMSE.RED r r.p SSE SSR
1 0.7091634 1.799126 0.4607073 0.8421184 5.346334e-55 188.2791 459.0917
SST Rsq stderr
1 647.3708 0.7091634 0.9702554mod$describe()Generalized Linear Model was used for regression.
R-squared was 0.71 (training).mod$plot()