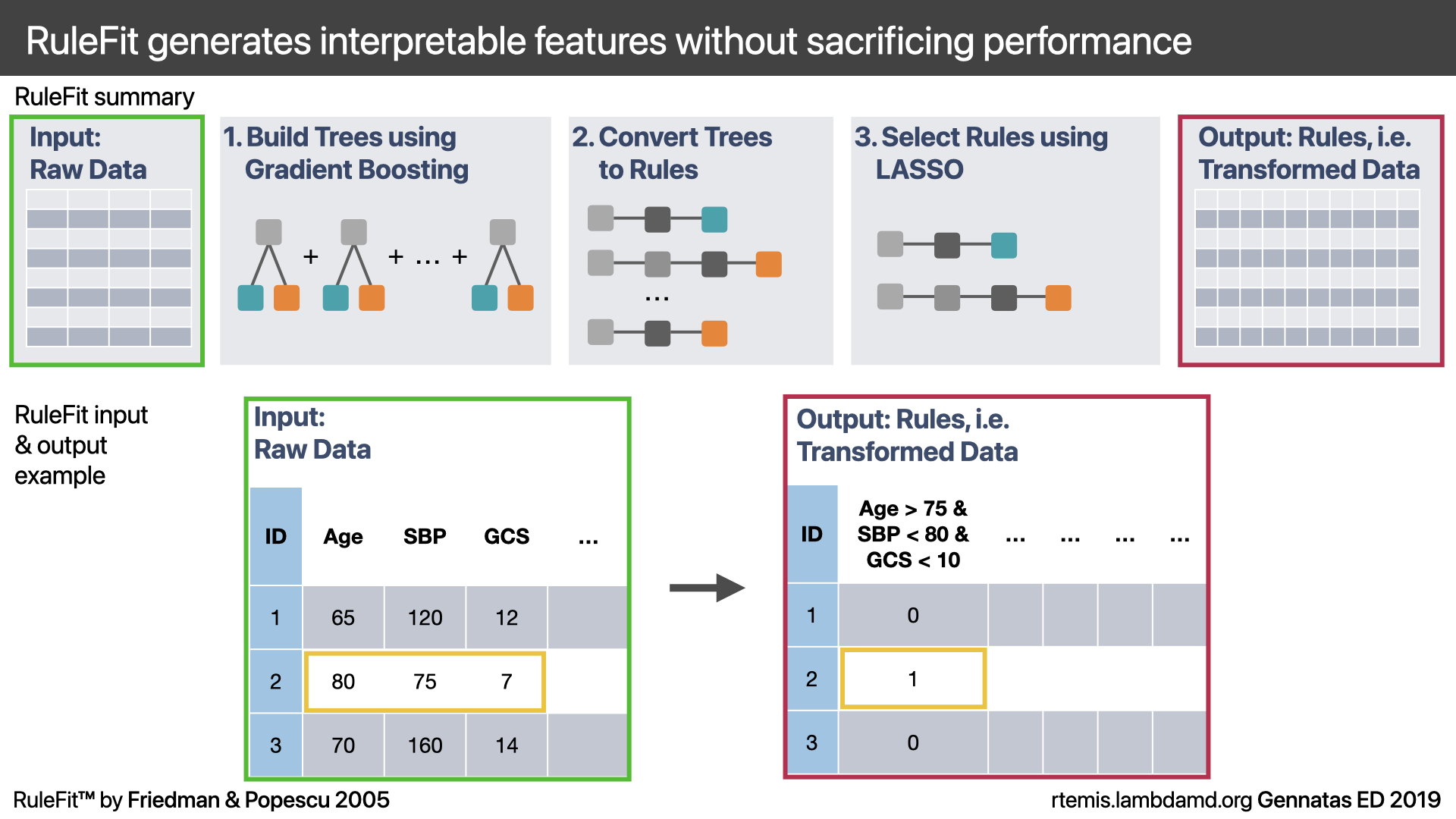
RuleFit is a powerful algorithm for regression and classification, which uses gradient boosting and the LASSO to train a highly accurate and interpretable model.
Given a dataset X and an outcome y:
Train a Gradient Boosting model on the raw inputs X to predict y
Take all decision tree base learners from (1) and convert them to a list of rules R (by following all paths from root node to leaf node). The rules represent a transformation of the raw input features.
Train a LASSO model on the ruleset R to predict y.
Thanks to the LASSO’s variable selection, step 3. will usually greatly reduce the large number of rules in R with no loss of accuracy. In fact, RuleFit may outperform gradient boosting.
Data
Let’s read in the Parkinsons dataset from the UCI repository:
<- read.csv ("https://archive.ics.uci.edu/ml/machine-learning-databases/parkinsons/parkinsons.data" )$ Status <- factor (parkinsons$ status, levels = c (1 , 0 ))$ status <- NULL $ name <- NULL check_data (parkinsons) parkinsons : A data.table with 195 rows and 23 columns
Data types
* 22 numeric features
* 0 integer features
* 1 factor, which is not ordered
* 0 character features
* 0 date features
Issues
* 0 constant features
* 0 duplicate cases
* 0 missing values
Recommendations
* Everything looks good
Resample
<- resample (parkinsons, seed = 2019 )06-30-24 10:57:35 Input contains more than one columns; will stratify on last [resample]
.: Resampling Parameters
n.resamples : 10
resampler : strat.sub
stratify.var : y
train.p : 0.75
strat.n.bins : 4
06-30-24 10:57:35 Using max n bins possible = 2 [strat.sub]
06-30-24 10:57:35 Created 10 stratified subsamples [resample]
<- parkinsons[res$ Subsample_1, ]<- parkinsons[- res$ Subsample_1, ]
RuleFit
<- s_RuleFit (park.train, park.test)06-30-24 10:57:35 Hello, egenn [s_RuleFit]
06-30-24 10:57:36 Running 1 Gradient Boosting Model... [s_RuleFit]
06-30-24 10:57:36 Hello, egenn [s_GBM]
06-30-24 10:57:36 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.: Classification Input Summary
Training features: 146 x 22
Training outcome: 146 x 1
Testing features: Not available
Testing outcome: Not available
06-30-24 10:57:36 Distribution set to bernoulli [s_GBM]
06-30-24 10:57:36 Running Gradient Boosting Classification with a bernoulli loss function [s_GBM]
.: Parameters
n.trees : 300
interaction.depth : 3
shrinkage : 0.1
bag.fraction : 1
n.minobsinnode : 5
weights : NULL
06-30-24 10:57:36 Training GBM on full training set... [s_GBM]
.: GBM Classification Training Summary
Reference
Estimated 1 0
1 110 0
0 0 36
Overall
Sensitivity 1.0000
Specificity 1.0000
Balanced Accuracy 1.0000
PPV 1.0000
NPV 1.0000
F1 1.0000
Accuracy 1.0000
AUC 1.0000
Brier Score 1.9e-08
Positive Class: 1
06-30-24 10:57:36 Completed in 8.8e-04 minutes (Real: 0.05; User: 0.05; System: 4e-03) [s_GBM]
06-30-24 10:57:36 Collecting Gradient Boosting Rules (Trees)... [s_RuleFit]
1200 rules (length<=3) were extracted from the first 300 trees.
06-30-24 10:57:36 Extracted 1200 rules... [s_RuleFit]
06-30-24 10:57:36 ...and kept 423 unique rules [s_RuleFit]
06-30-24 10:57:36 Matching 423 rules to 146 cases... ✓ [matchCasesByRules]
06-30-24 10:57:36 Running LASSO on GBM rules... [s_RuleFit]
06-30-24 10:57:36 Hello, egenn
06-30-24 10:57:36 Imbalanced classes: using Inverse Frequency Weighting [prepare_data]
.: Classification Input Summary
Training features: 146 x 423
Training outcome: 146 x 1
Testing features: Not available
Testing outcome: Not available
06-30-24 10:57:36 Running grid search... [gridSearchLearn]
.: Resampling Parameters
n.resamples : 5
resampler : kfold
stratify.var : y
strat.n.bins : 4
06-30-24 10:57:36 Using max n bins possible = 2 [kfold]
06-30-24 10:57:36 Created 5 independent folds [resample]
.: Search parameters
grid.params :
alpha : 1
fixed.params :
.gs : TRUE
which.cv.lambda : lambda.1se
06-30-24 10:57:36 Tuning Elastic Net by exhaustive grid search. [gridSearchLearn]
06-30-24 10:57:36 5 inner resamples; 5 models total; running on 8 workers (aarch64-apple-darwin20) [gridSearchLearn]
06-30-24 10:57:37 Extracting best lambda from GLMNET models... [gridSearchLearn]
.: Best parameters to maximize Balanced Accuracy
best.tune :
lambda : 0.0370390609993396
alpha : 1
06-30-24 10:57:37 Completed in 0.01 minutes (Real: 0.62; User: 0.09; System: 0.08) [gridSearchLearn]
.: Parameters
alpha : 1
lambda : 0.0370390609993396
06-30-24 10:57:37 Training elastic net model...
.: GLMNET Classification Training Summary
Reference
Estimated 1 0
1 110 0
0 0 36
Overall
Sensitivity 1.0000
Specificity 1.0000
Balanced Accuracy 1.0000
PPV 1.0000
NPV 1.0000
F1 1.0000
Accuracy 1.0000
AUC 1.0000
Brier Score 0.0106
Positive Class: 1
06-30-24 10:57:37 Completed in 0.01 minutes (Real: 0.78; User: 0.23; System: 0.10)
.: RuleFit Classification Training Summary
Reference
Estimated 1 0
1 110 0
0 0 36
Overall
Sensitivity 1.0000
Specificity 1.0000
Balanced Accuracy 1.0000
PPV 1.0000
NPV 1.0000
F1 1.0000
Accuracy 1.0000
AUC 1.0000
Brier Score 0.0106
Positive Class: 1
06-30-24 10:57:37 Matching cases to rules... [predict.rulefit]
06-30-24 10:57:37 Matching 423 rules to 49 cases... ✓ [matchCasesByRules]
.: RuleFit Classification Testing Summary
Reference
Estimated 1 0
1 34 3
0 3 9
Overall
Sensitivity 0.9189
Specificity 0.7500
Balanced Accuracy 0.8345
PPV 0.9189
NPV 0.7500
F1 0.9189
Accuracy 0.8776
AUC 0.9212
Brier Score 0.0966
Positive Class: 1
06-30-24 10:57:37 Completed in 0.03 minutes (Real: 1.54; User: 0.95; System: 0.12) [s_RuleFit]
RuleFeat Output
Let’s explore the algorithm output. The rules with their associated coefficients and empirical risk are stored in park.rf$mod$rules.selected.coef.er:
R-readable rules
We can also access the R-readable rules directly:
$ mod$ rules.selected
[1] "MDVP.Fo.Hz.>133.131 & MDVP.Fhi.Hz.<=204.673 & PPE<=0.184526"
[2] "D2>2.0310275 & PPE>0.1345545"
[3] "D2>2.0459605 & PPE>0.150139"
[4] "MDVP.Fo.Hz.<=208.8315 & D2>2.224767"
[5] "MDVP.Fhi.Hz.<=205.209 & spread2<=0.193501 & PPE<=0.1643355"
[6] "MDVP.Fhi.Hz.>205.209 & spread2<=0.193501"
[7] "MDVP.Flo.Hz.>77.8015 & Shimmer.APQ5>0.006365 & spread1<=-5.618097"
[8] "Shimmer.APQ3>0.00745 & MDVP.APQ<=0.019775"
[9] "MDVP.Fo.Hz.<=208.8315 & NHR>0.004855 & RPDE>0.339766"
[10] "DFA>0.6888735 & DFA<=0.7325515 & spread1<=-5.618097"
[11] "MDVP.Fhi.Hz.<=229.1795 & NHR>0.004855 & RPDE>0.339766"
[12] "MDVP.Fo.Hz.>117.548 & MDVP.Shimmer.dB.<=0.1875 & PPE>0.1345545"
[13] "Shimmer.APQ3>0.00745 & PPE<=0.184526"
[14] "MDVP.Fo.Hz.>117.548 & MDVP.Shimmer.dB.<=0.1895 & PPE>0.1345545"
[15] "MDVP.Fhi.Hz.<=229.1795 & MDVP.RAP>0.001885 & RPDE>0.339766"
[16] "MDVP.Fo.Hz.<=133.131 & HNR<=26.804 & spread1<=-5.618097"
[17] "MDVP.Fhi.Hz.<=204.673 & MDVP.Jitter...<=0.003315 & PPE<=0.184526"
[18] "Shimmer.APQ3>0.00843 & Shimmer.APQ3>0.008815 & spread1>-6.4767615"
[19] "MDVP.Fhi.Hz.<=206.449 & MDVP.Jitter...<=0.003315"
[20] "MDVP.Fo.Hz.<=118.7465 & MDVP.Jitter...<=0.00449 & D2<=2.0310275"
[21] "RPDE>0.367432 & PPE<=0.184526"
[22] "MDVP.Jitter...<=0.00449 & D2<=2.0310275"
[23] "MDVP.Jitter...>0.00411 & Jitter.DDP<=0.00563"
[24] "MDVP.Fo.Hz.>118.3085 & MDVP.APQ<=0.013695 & spread1>-6.512704"